FIG 1.
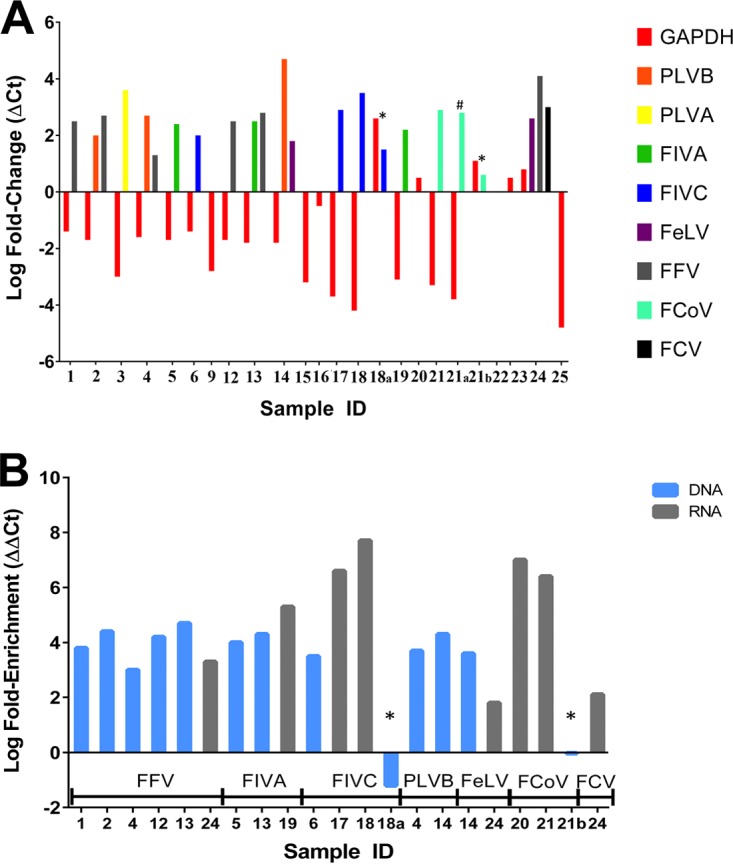
(A) Targeted genome capture results in the enrichment and depletion of pathogen and host nucleic acids, respectively. RT-PCR was performed pre- and postcapture for feline genomic DNA/cDNA (GAPDH) and individual pathogens pre- and postcapture. Two unenriched samples (*) demonstrated an increase of ≥1-fold in the relative abundance of the host GAPDH gene, compared to all of the enriched samples tested, in which GAPDH either decreased or remained approximately the same. One sample (#) was enriched with only half of the recommended capture probes but still demonstrated a depletion of host nucleic acids and an increase in pathogen nucleic acids. All pathogen nucleic acids measured increased in relative abundance postenrichment. Pathogen and host nucleic acids were measured only in samples with sufficient volume remaining before and after sequencing to conduct RT-PCRs. (B) Pathogen nucleic acids were enriched from 58-fold to 56 million-fold relative to host nucleic acids. The relative abundance of pathogen DNA did not increase in the two unenriched samples (*), but the corresponding paired, enriched pathogens increased over 1 million-fold. Samples in this figure are grouped by pathogen type: FFV, feline foamy virus; FIVA, feline immunodeficiency virus clade A; FIVC, feline immunodeficiency virus clade C; PLVB, puma lentivirus clade B; FeLV, feline leukemia virus; FCoV, feline corona virus; FCV, feline calicivirus.
