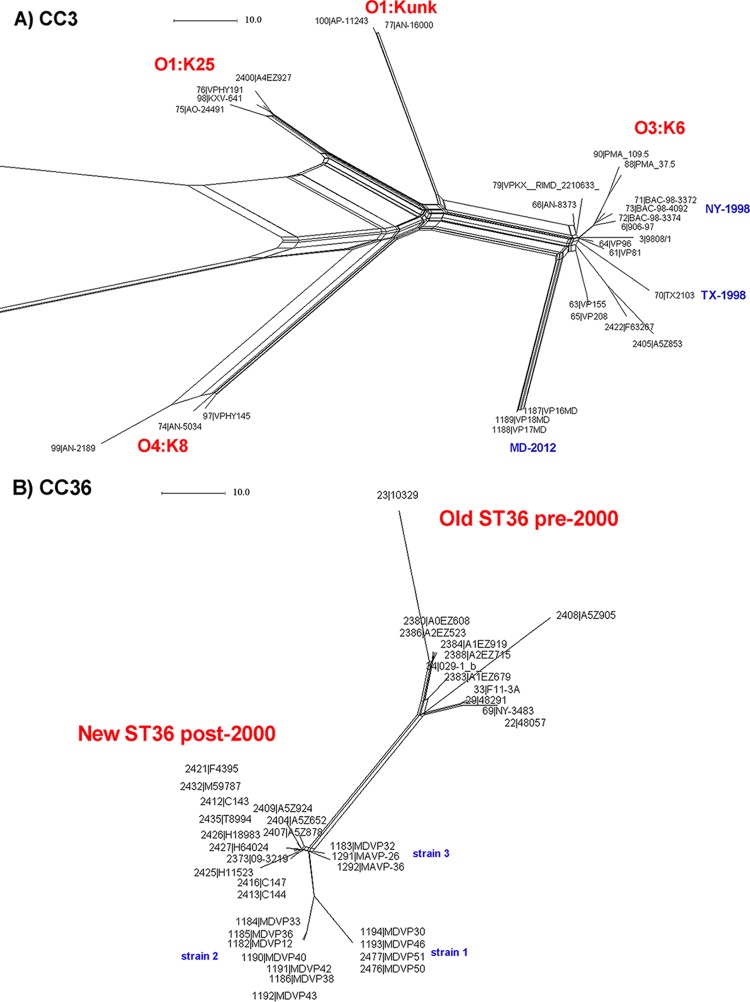
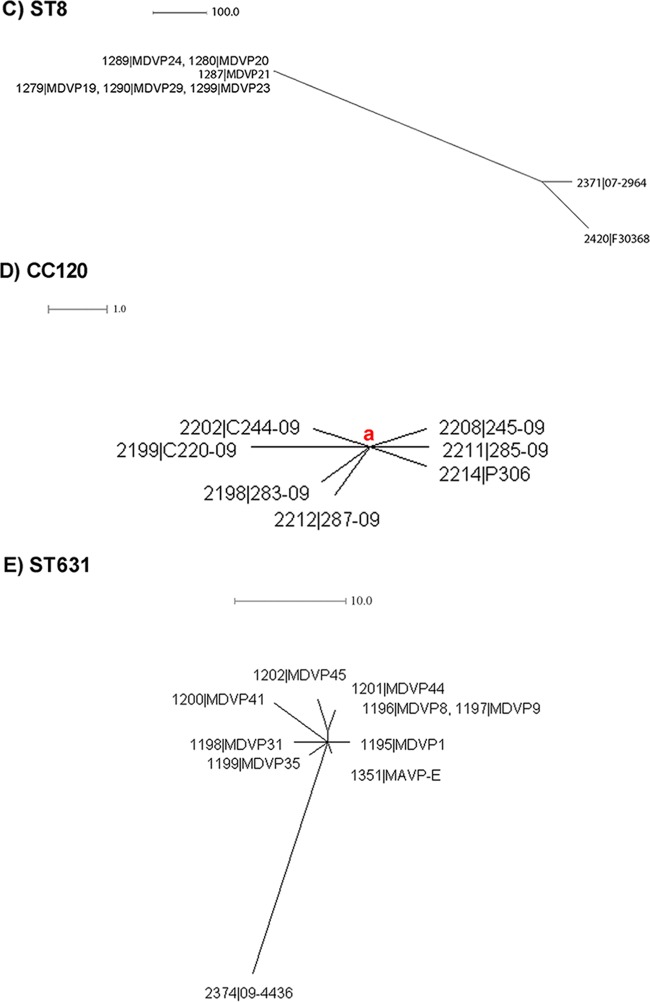
FIG 2.
cgMLST analysis of representative V. parahaemolyticus strains from same outbreaks and/or non-outbreak related displaying the same ST identified in Fig. 1. (A) CC3 outbreak-related (12) and non-outbreak-related (19). (B) CC36-ST36 outbreak-related and non-outbreak-related strains (5, 19, 36). (C) CC8-ST8 outbreak-related and non-outbreak-related strains (15). (D) CC120 outbreak (Peru, 2009 [16]) strains 281-09, 241-09, 379-09, CO1409, CO1609, P310, Guillen_151_Peru, C226-09, C224-09, C235, PIURA_17, C237, and 239-09, were identical by cgMLST (represented by letter a). (E) ST631 strains (5, 34, 36). The scale represents the number of allele differences.