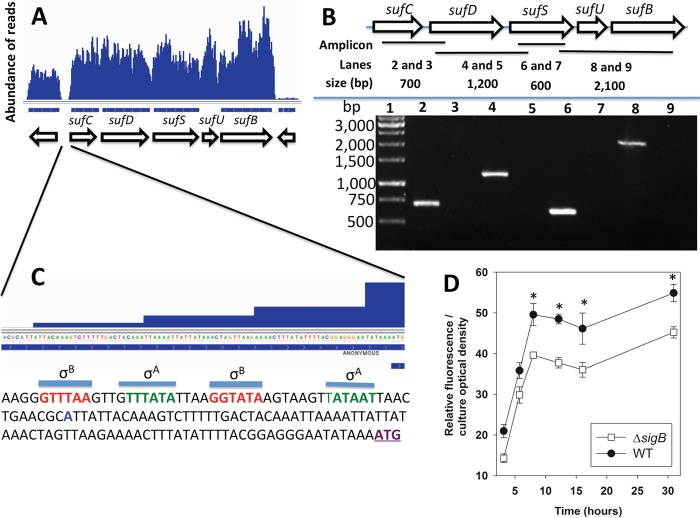
FIG 3.
The sufCDSUB genes are cotranscribed, and transcription is positively influenced by sigma factor B. (A) Analysis of a previously published RNA-seq data set (29) indicating that sufCDSUB are cotranscribed. (B) The suf genes are cotranscribed. (Top) Schematic of the suf operon; the locations of the amplicons are shown as black bars, and the predicted sizes of the amplicons (generated using the following primer pairs: lanes 2 and 3, sufCRT5 and sufDrevRT; lanes 4 and 5, sufDfwdRT and sufinternal3; lanes 6 and 7, sufinternal5 and sufSrevRT; lanes 8 and 9, sufSfwdRT and sufBrevRT) are shown. (Bottom) Amplicons were generated from cDNA libraries using RNAs isolated from the WT and separated using agarose gel electrophoresis. The samples analyzed in lanes 3, 5, 7, and 9 were generated using a template that was not treated with reverse transcriptase. (C) The promoter of the suf operon contains potential sigma factor A (green) and sigma factor B (red) recognition sites. The predicted transcriptional start site is shown in blue and was determined by analyzing previously published RNA-seq data (29). The annotated sufC translational start site is in purple and underlined. (D) The transcriptional activity of the sufC promoter is modulated by sigma factor B (SigB). The transcriptional activity of sufC was monitored in the WT and ΔsigB strains containing pCM11_suf. The data shown represent the averages of biological triplicates with standard deviations. *, P < 0.5 relative to the WT strain, using a two-tailed Student t test.