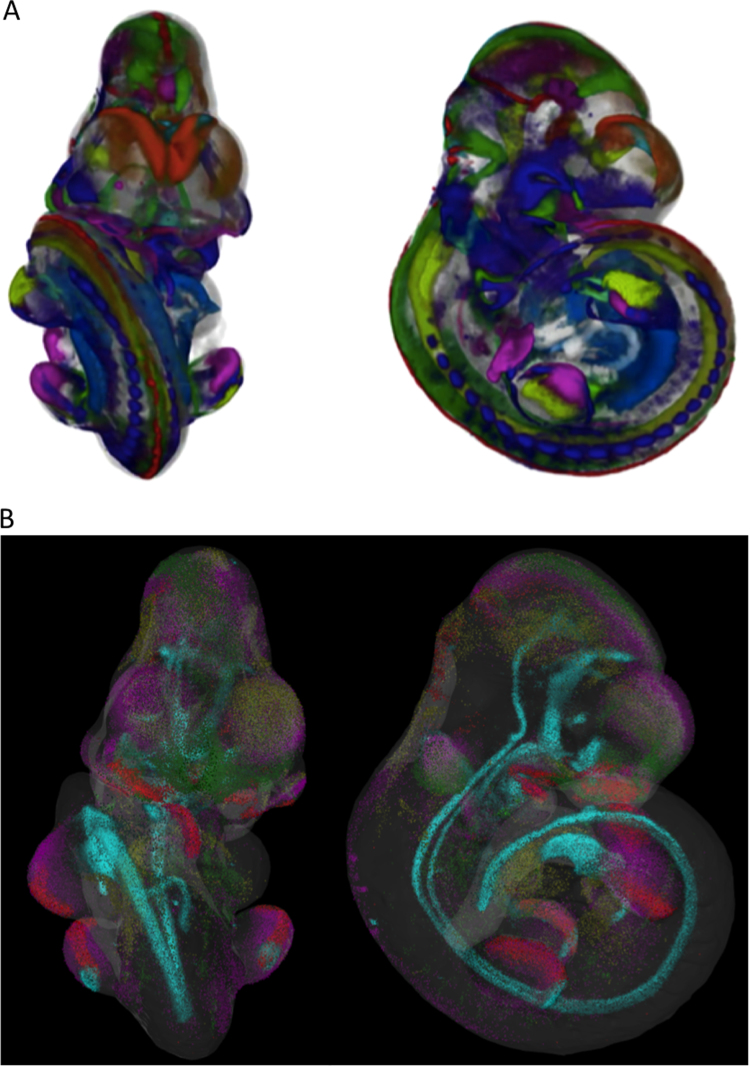
Fig. 2.
3D visualisation of molecular anatomy. A) Volume rendered views of 19 E10.5 (TS17) spatially mapped Wnt family gene expression patterns can be generated using commercially available software packages such as Amira. These visualisations can be used to cross-compare gene expression patterns in a spatial framework. B) Point-cloud rendered views of spatially mapped E10.5 (TS17) Shh (cyan), Fgf8 (red), Fgf9 (green), Fgf10 (magenta), and Fgf20 (yellow) enable direct visualisation of spatially mapped gene expression patterns in the context of a web browser and without the need for data download. An advantage of the point cloud visualisation is that a user can see through the entire 3D volume, and can more accurately identify regions of expression. An interactive point cloud visualisation can be found at the following link: http://aberlour.hgu.mrc.ac.uk/MARenderTests/genexpr-shh+fgf.html.