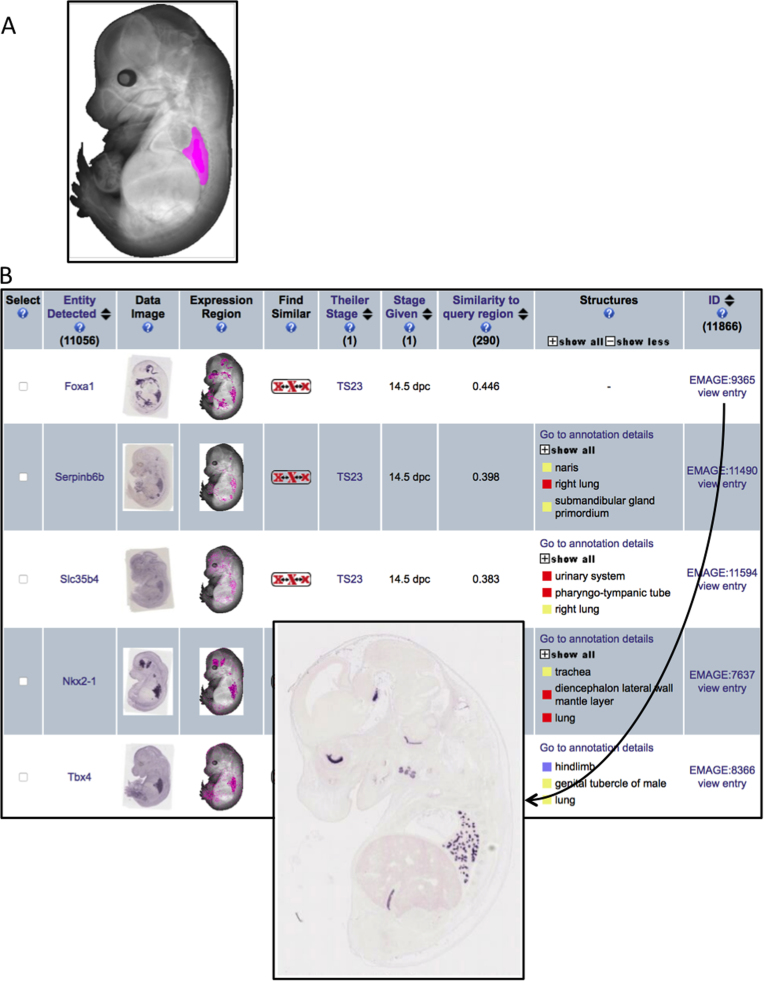
Fig. 3.
EMAGE Spatial Query. A use case scenario in which an EMAGE spatial query is used to identify gene expression patterns in the developing lung. A) A user-defined region (pink) on an embryo model is delineated using the Embryo Space paint query tool. B) A results table returns a list of EMAGE entries ranked by spatial similarity to the defined region of interest. The gene symbol (entity detected), spatial annotation (expression region), text annotation (structures), and similarity score (similarity to query region) are all shown in the results table. High-resolution original images (inset) and probe details are found on the EMAGE entry page, which can be accessed by clicking on the EMAGE entry ID in the results table. This example query identified Foxa1, Serpin6b, Slc35b4, Nkx2-1, and Tbx4 as the top 5 genes that show spatially mapped expression in the E14.5 (TS23) lung. The first gene in this list, Foxa1, was not text-annotated by the authors but was spatially mapped by the EMAGE Editorial Office and so could be retrieved by spatial query.