Fig. 1.
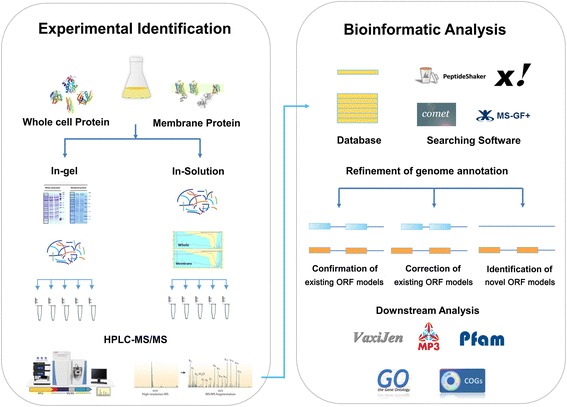
Experimental identification and bioinformatics analysis workflow of the proteogenomics study. Whole cell protein and membrane protein extracts were prepared from B. abortus 104 M cultures grown to exponential phase. Complexity reduction was achieved by SDS-PAGE and SCX HPLC separation. After protein extraction and pre-fractionation, enzymatically digested proteins or peptides were analyzed by LC-MS/MS. All spectra were searched against the six-frame (6 F) database and the UniProt database by SearchGUI. Identified peptides were mapped to the 104 M genome and annotation was refined at three levels: (i) confirmation of the existing open reading frame (ORF) models; (ii) refinement of the existing ORF models; and (iii) identification of novel ORF models. Subsequently, the protein physical\chemical property and function were analyzed
