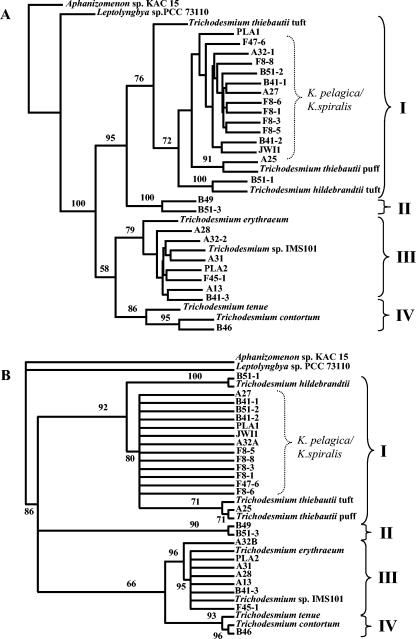
FIG. 3.
Phylogenetic trees inferred from partial hetR sequences obtained in the present study from natural and cultured populations of Katagnymene and Trichodesmium species and from the extant Trichodesmium cluster (14). Aphanizomenon sp. strain KAC15 and Leptolyngbya sp. strain PCC 73110 were used as outgroups. The brackets with roman numbers indicate the four main clusters in the Trichodesmium cluster. The brackets labeled K. pelagica/K. spiralis indicate the 13 sequences derived from trichomes originally identified as K. pelagica or K. spiralis. (A) Maximum-likelihood analysis of partial hetR nucleotide sequences. (B) Maximum-likelihood analysis of partial hetR amino acid sequences. For the analyses of the amino acid sequences encoded by hetR, TREE-PUZZLE 5.0 (27) was performed with the default settings, except that the model of rate heterogeneity was chosen as a gamma distributed rate. Abbreviations: IMS, culture collection of the Institute of Marine Science, University of North Carolina, Morehead City; KAC, Kalmar Algae Collection; PCC, Pasteur Culture Collection.