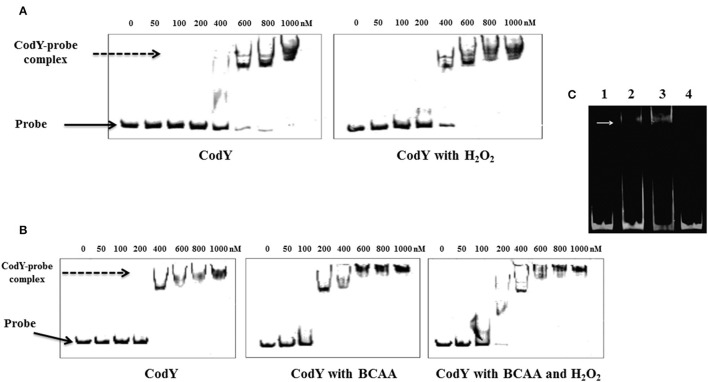
Figure 4.
CodY interaction with the regulatory region of tpxD. EMSA assay was used to confirm the presence of CodY binding site in tpxD regulatory region: Effect of H2O2 (A) and BCAA (B). 10 nM of fluorescently labeled DNA probe was mixed with increasing concentration of recombinant CodY, and incubated for 30 min at room temperature. When required, 2 mM H2O2 and/or BCAA (isoleucine 1.62 mM, leucine 3.48 mM, and valine 2.77 mM) were added into the binding buffer. DNA-protein complexes were separated by electrophoresis on non-denaturing PAGE (8%). In all gels, Lane 1 contains DNA probe alone, indicated with a black arrow. DNA-protein interaction is seen as an upward shift of DNA probe, indicated with a dotted arrow. (C) Lane 1, 50 ng approximately of the 81 bp DNA fragment alone, lane 2 and 3, DNA plus 0.6- and 1-mM recombinant CodY, respectively. Lane 4, DNA probe, excluding 15 bp putative CodY binding site plus 1 mM recombinant CodY. White arrow indicates the position of DNA-protein complex. EMSA was performed using Molecular Probes fluorescence-based EMSA kit (Invitrogen). Briefly, 5X FY binding buffer (20 mM Tris-HCl pH 7.5, 30 mM KCl, 1 mM DTT, 1 mM EDTA pH 8.0 and 10% v/v glycerol) was prepared to incubate the promoter probe and protein. The binding reaction was set up by mixing a constant amount of target promoter probe (~30 ng), and increasing amounts (0.1–0.5 μM) of purified and dialyzed His-tagged protein. The binding reaction was incubated at room temperature for 20 min in a total volume of 20 μl, and then analyzed on an 8% w/v non-denaturing polyacrylamide gel. After electrophoresis, gels were stained with SYBR® Green EMSA gel stain (Invitrogen) and visualized using a Typhoon Trio+ scanner (GE Healthcare Life Sciences) with a 526 nm short-pass wavelength filter.