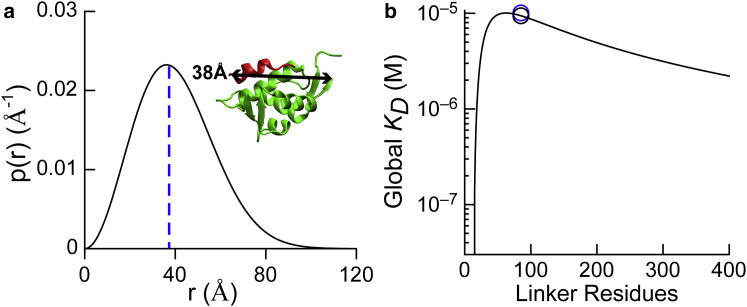
Figure 2.
Modeling MdmX intramolecular interactions using polymer statistics. (a) Shown here is the probability distribution of end-to-end distances for the MdmX linker region (residues 110–194). The distribution peaks at 36 Å are given. The blue dashed line marks 38 Å, the measured distance between the end of the MdmX domain and the beginning of the WW motif considering the same binding orientation as p53TAD, measured from PDB: 3DAB. The ribbon model shows a structure from PDB: 3DAB. (b) Shown here is the predicted global KD as a function of linker length for a direct binding orientation model. Black circle: global KD predicted by the WLC model for an 85-residue linker; blue circle: experimentally measured global KD (see Fig. 3).