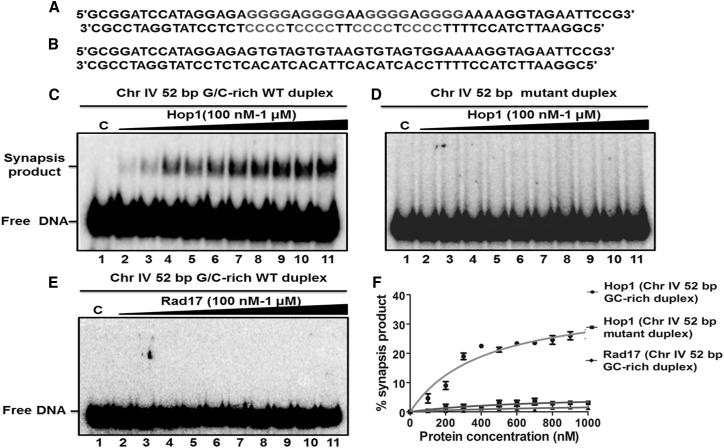
Figure 11.
The pairing of double-stranded DNA by Hop1 is dependent on the presence of a G/C-rich sequence. (A) Given here is a schematic representation of the 52-bp duplex DNA containing the G/C-rich motif embedded at the center. (B) Given here is a schematic representation of duplex DNA with a mixed sequence. (C) DNA pairing assays were carried out with a 52-bp G/C-rich duplex DNA with increasing concentrations of Hop1. (D) The pairing of duplex DNA molecules with a mixed sequence in the presence of increasing concentrations of Hop1 is shown. Reaction mixtures (20 μL) contained 2 nM of the indicated 32P-labeled duplex DNA in the absence (lane 1) or presence of 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 1 μM of Hop1(lanes 2–11), respectively. (E) DNA pairing assays were performed with a 52-bp duplex DNA in the absence (lane 1) and in the presence of 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 1 μM Rad17 (lanes 2–11), respectively. The samples were incubated with proteinase K and SDS and analyzed as described under Materials and Methods. The positions of free DNA and products are indicated on the left-hand side of the image. Lane 1 was the reaction performed in the absence of protein. (F) Given here is a graphical representation of the extent of the generation of a synapsis product with G/C-rich duplex DNA or mutant duplex DNA. The extent of the product formed in (C)–(E) is plotted versus varying concentrations of the specified protein. Error bars indicate mean ± SE.