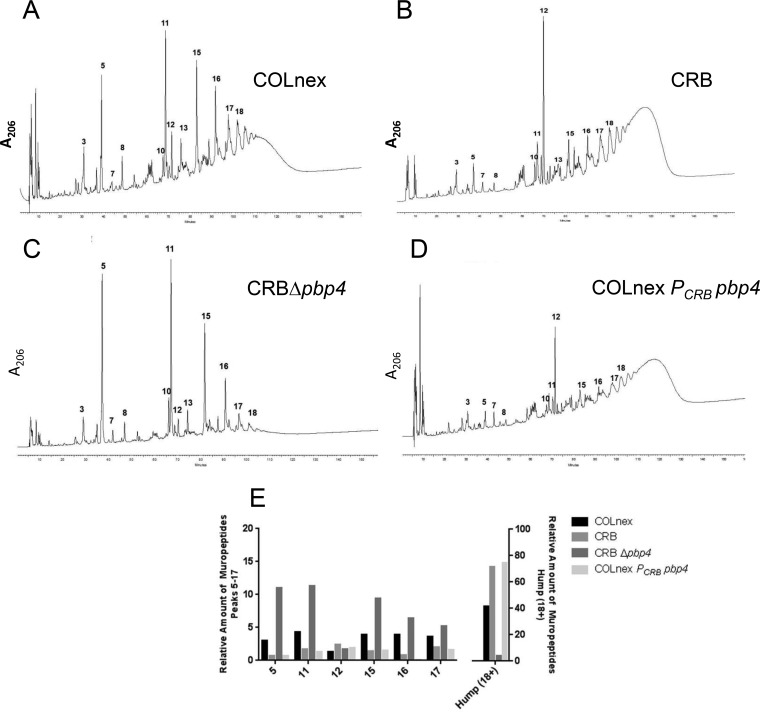
FIG 3.
Changes in the peptidoglycan compositions of S. aureus CRB mutant strains. (A to D) Peptidoglycans were prepared, and muropeptide patterns were analyzed by HPLC for the COLnex parental strain (A), mutant CRB (B), CRB in which pbp4 was deleted (CRB Δpbp4) (C), and the COLnex PCRB pbp4 strain with the CRB promoter mutation (D). Peaks 5, 11, 15, 16, and 17 represent the mono-, di-, tri-, tetra-, and pentameric muropeptide derivatives, respectively. Peaks 18+ (the hump) contain highly cross-linked oligomeric components. (E) Quantitative differences in the peptidoglycan compositions of COLnex, CRB, and their derivatives. The bars show the relative amounts of muropeptides calculated from the UV absorbance of the peaks in relation to the total amount of muropeptides. The relative amounts of muropeptides for peaks 5 to 17 and 18+ (the hump) are depicted on the left and right axes, respectively.