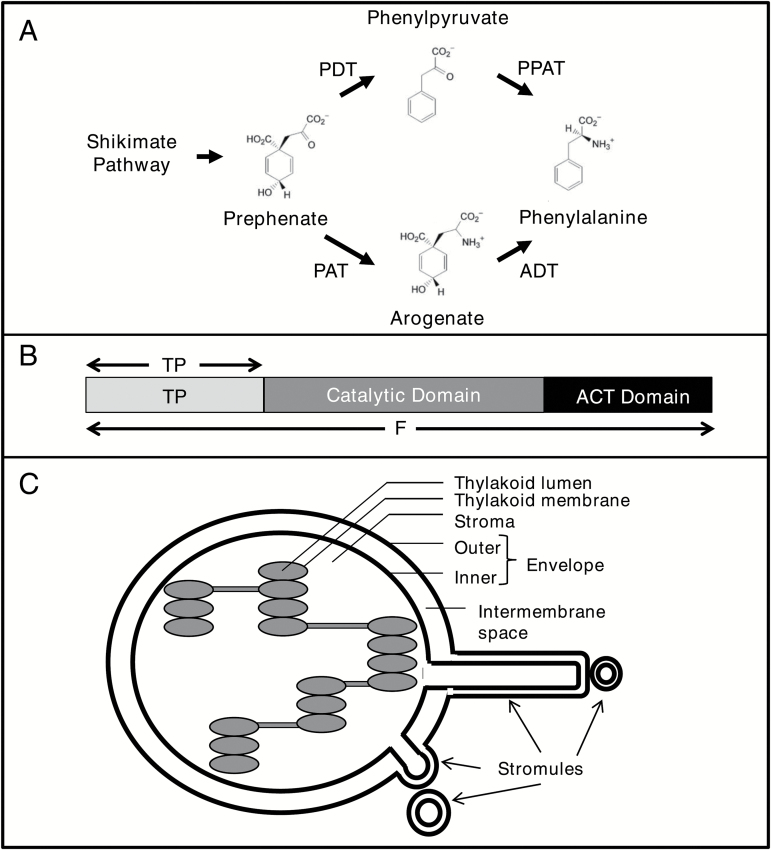
Fig. 1.
Phenylalanine synthesis, arogenate dehydratases, and stromules. (A) Phenylalanine can be synthesized in plants using either the prephenate (top) or the arogenate (bottom) pathway (Cho et al., 2007; Maeda and Dudareva, 2012). Prephenate is either decarboxylated/dehydrated to phenylpyruvate (PP) by a prephenate dehydratase (PDT) and PP is then transaminated by a phenylpyruvate aminotransferase (PPAT) to phenylalanine. Alternatively the two enzymatic steps are reversed, whereby prephenate is transaminated to arogenate by a prephenate aminotransferase (PAT) and arogenate is then decarboxylated/dehydrated to phenylalanine by an arogenate dehydratase (ADT). (B) A. thaliana ADT constructs were cloned in different lengths. The full-length (F) sequence represents the entire ADT ORF while an N-terminal construct only includes the transit peptide (TP). (C) Schematic diagram of a chloroplast showing the formation of stromules. Stromules are stroma-filled protrusions of the outer and inner membrane from chloroplasts. They can differ in length, forming long thread-like extensions or globular structures.