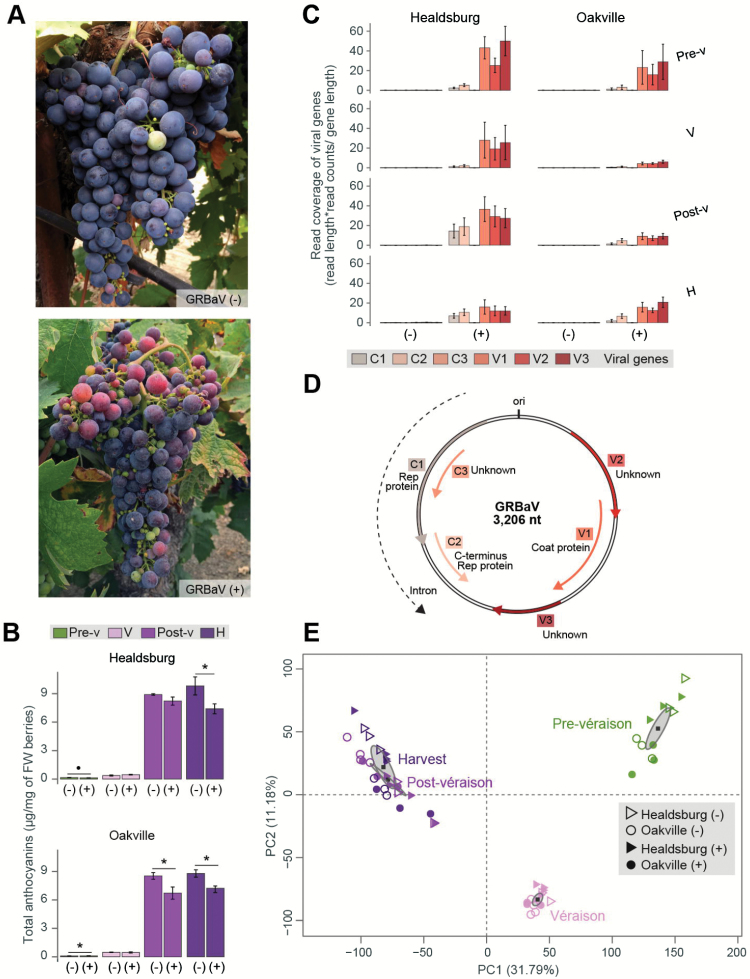
Fig. 1.
Naturally occurring GRBaV infections in a red-skinned grapevine cultivar. (A) Berry cluster asynchronous development observed in grapevines that tested positive for the presence of GRBaV. (B) Total anthocyanins from GRBaV-positive and GRBaV-negative berries collected at four specific ripening stages. Symbols indicate significant fold changes (•P≤0.1, *P≤0.05). (C) Expression of GBRaV genes in berries from GRBaV-positive (+) or negative (–) control vines across four ripening stages represented as mapped read coverage based on RNAseq data. (D) Genome organization of GRBaV obtained from Krenz et al. (2014). Six ORFs are denoted with colored arrows and their gene names are included. The origin of replication (ori) is also depicted. (E) Principal component analysis of GRBaV-positive and negative berries at four ripening stages and collected from two independent vineyards. The quantitative variables correspond to transcript abundance of 25 995 grape genes. Each circle or triangle represents a biological replicate. Gray ellipses define confidence areas (95%) for each stage, while gray squares represent their corresponding centers of gravity. Pre-v, pre-véraison; V, véraison; Post-v, post-véraison; H, harvest; PC, principal component. Error bars correspond to SEs.