Fig. 2.
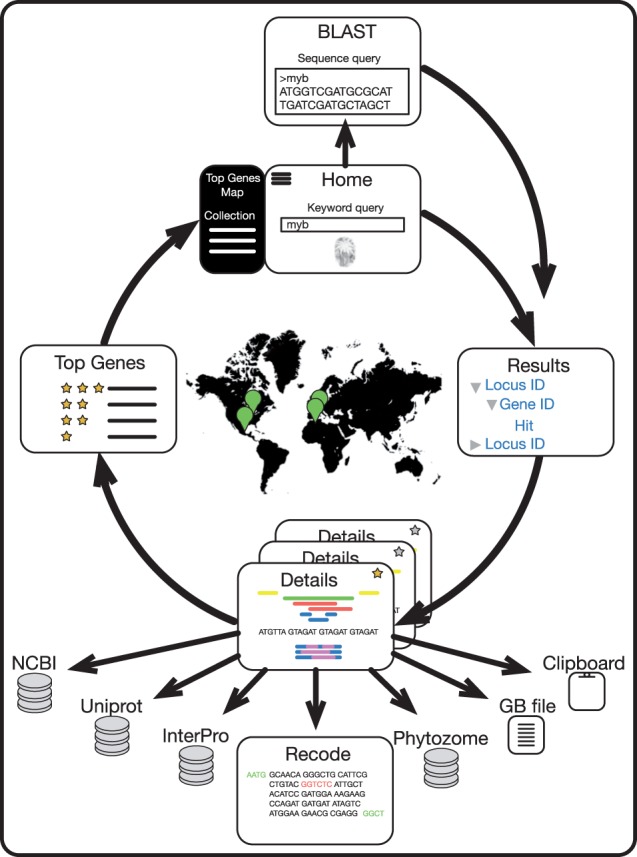
MarpoDB workflow. Top: Upon query submission at the Home or BLAST page, a user is presented with a table of results (right), where each row corresponds to a gene entry, containing a query keyword. Bottom: Selecting one of the results from the list opens up the Details page, which contains the gene model chart, and sequence viewer along with BLAST and Pfam annotations. From the Details page, the user can proceed with a BLAST search of the selected sequence in the NCBI database; export the gene model in GenBank format; or copy the selected sequence into the clipboard. Alternatively the user may proceed to the Recode page for adapting the selected sequence for type IIS assembly. In addition, the user can ‘star’ a gene, which will add it to the user’s personal collection, as well as increase the gene score in the global top (left).
