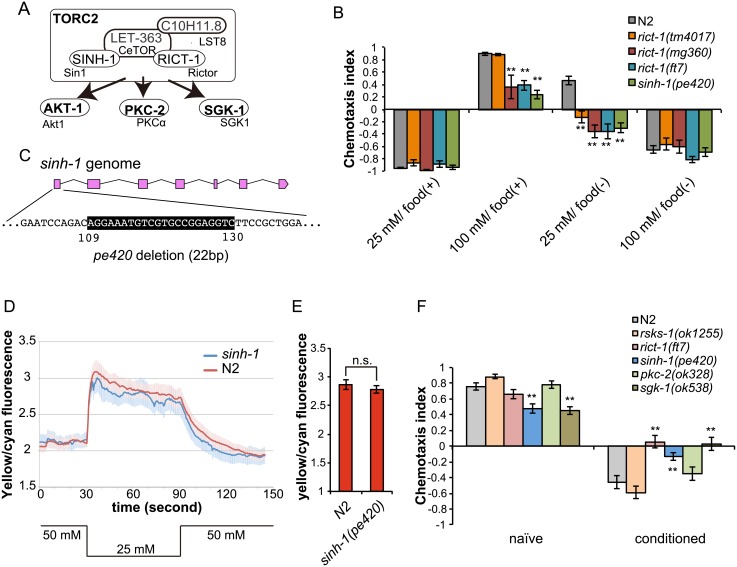
Fig 3. TORC2 components regulate migration to high salt concentrations.
(A) A schematic model of putative TORC2 signaling in C. elegans. (B) Salt chemotaxis of wild-type N2 worms and mutants of TORC2 components. TORC2-signaling mutants migrated to lower salt levels than did wild-type worms. (C) Genomic structure of sinh-1. Pink boxes indicate exons. The location of the pe420 deletion is shown below the sinh-1 locus. (D) Calcium responses of ASER of wild-type animals and sinh-1 mutants to an NaCl down-step from 50 to 25 mM and back to 50 mM (traces represent the means, the shadings the s.e.m.) after 5-h low-salt/food(–) conditioning. (E) Bar graphs showing averaged yellow/cyan fluorescence ratios during 10 s after downstep stimulation. Error bars, s.e.m.; n.s. = not significant (Student’s t test, N ≥ 9). (F) Benzaldehyde chemotaxis of wild-type N2 and mutant worms after benzaldehyde conditioning or in the naive state. rict-1(ft7), sinh-1(pe420), and sgk-1(ok538) mutants, but not pkc-2(ok328) and rsks-1(ok1255) mutants, showed defects in food-odor associative learning. Error bars, s.e.m.; **p < 0.01, n.s. = not significant (wild-type vs. each mutant, one-way ANOVA with Dunnett’s post hoc test, N ≥ 9).