Figure 3.
Identification of the CPT1 Gene.
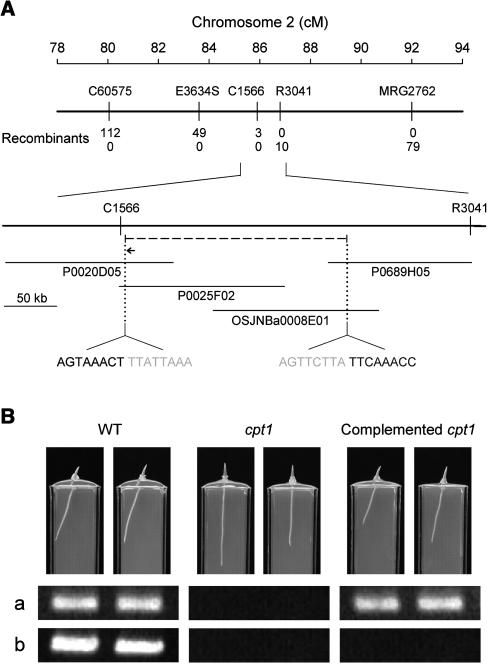
(A) Map-based cloning. The CPT1 locus was mapped to chromosome 2. C60575 and MRG2762 are STS markers. E3634S, C1566, and R3041 are RFLP markers. Numbers of recombinants between cpt1 and each marker are shown. P0020D05, P0025F02, and P0689H05 are PAC clones, and OSJNBa0008E01 is a BAC clone. The cpt1 mutant was found to have a deletion of ∼209 kb (dashed line) between C1566 and R3041 markers that enclosed the CPT1 locus. Sequences around the two edges of the deletion are indicated (gray letters represent deletion). Within the deletion, a NPH3-like gene (indicated by a horizontal arrow; 4.5 kb) was predicted. cM, centimorgan.
(B) Transgenic complementation of the cpt1 mutant. A genomic clone (8.2 kb) containing the NPH3-like gene was introduced into the cpt1 mutant by Agrobacterium-mediated transformation. The photographs show 3-d-old seedlings of the wild type, cpt1 mutant, and complemented cpt1 mutant (T3 generation) grown under irradiation with unilateral white light (3 μmol m−2 s−1; from the right side of the photographs) and overhead red light (3 μmol m−2 s−1). The two complemented cpt1 seedlings originated from independent transformants. Bottom panels show ethidium bromide–stained PCR products obtained for the photographed plants. (a) A 428-bp fragment of the NPH3-like gene was amplified with primers specific to this gene (5′-TTGCAGTGCATAGCCAGTAC-3′ and 5′-TTTCCACGTACTTCTCGTCC-3′). (b) A 502-bp genomic fragment located in the deleted region and at a close distance from the NPH3-like gene was amplified (primers 5′-CGTTGTACGACTGGATGGAC-3′ and 5′-CACTCAACCTTCCGCTTCTC-3′).