FIGURE 5.
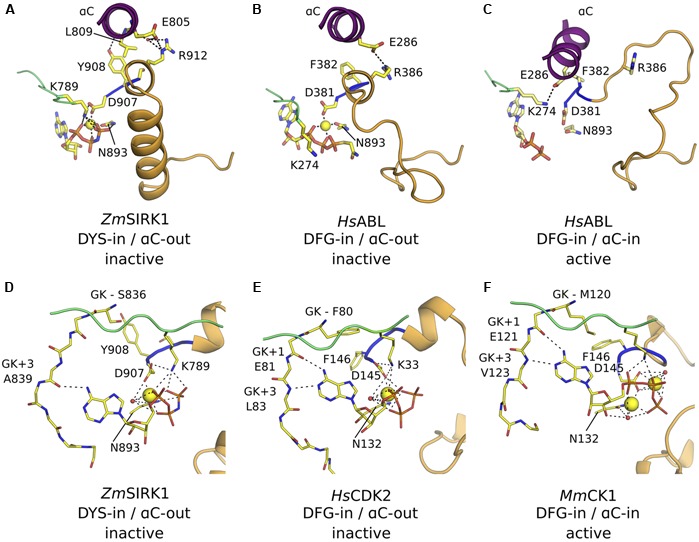
ZmSIRK1 adopts an inactive DFG-in/αC-out conformation. (A) Cartoon representation of ZmSIRK1 around αC, DYS motif and activation segment showing residues (stick) and interactions (dashed lines) that stabilize the inactive conformation in this protein. (B) Human proteins, such as HsABL (PDB ID 2G1T), can also adopt the DFG-in/αC-out conformation. (C) Phosphorylation of regulatory sites (not shown) within HsABL stabilizes the active (DFG-in/αC-in) conformation. (D) Details of ZmSIRK1 interaction with Mg2+ ion and AMP-PNP showing the unusual Mg2+ ion-coordination, ribose puckering and adenine ring interactions in ZmSIRK1-AMP-PNP co-structure. Possible hydrogen bonds are shown as dashes. (E) Inactive state human proteins, such as CDK2 (PDB ID 1HCK) can also display unusual Mg2+ ion coordination and sugar puckering. (F) Mg2+ ion coordination and nucleotide interaction for the prototypical active state kinase domain of Mus musculus (Ms)CK1 (PDB ID 1ATP). Residues in the hinge region are annotated relative to the gatekeeper residue (GK, GK+1, GK+3).
