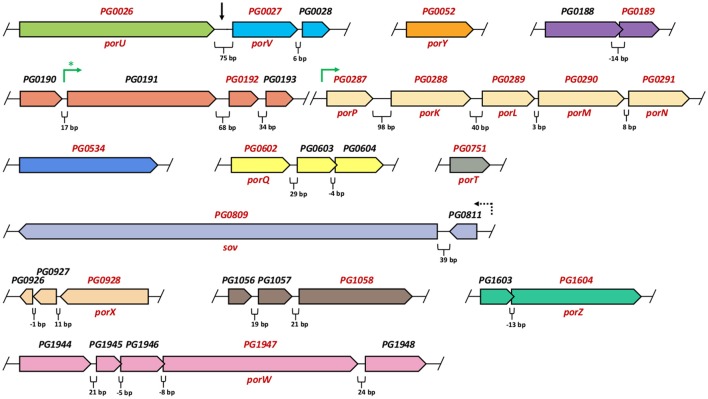
Figure 2.
Arrangement of P. gingivalis W83 genes encoding T9SS components. Genes are grouped according to in silico operon predictions, reflecting direction of transcripts (Dam et al., 2007; Mao et al., 2009; Pertea et al., 2009; Taboada et al., 2012). Gaps in the genome are indicated by the slashes. Intervals between adjacent genes or overlapping regions (in base pairs-bp) are marked below each section. Each transcription unit is shown in different color. Genes encoding T9SS components are depicted in red font. Black vertical arrow shows continuous region (75 bp) between PG0026 (porU) and PG0027 (porV) but the two genes were predicted to transcribe independently. Green arrows indicate operons that were confirmed experimentally (Taguchi et al., 2015; Vincent et al., 2017). Green asterisk denotes proved single transcription unit for the PG0191-PG0192-PG0193 genes (in P. gingivalis ATCC33277 strain), however co-transcription of preceding the PG0190 gene (17 bp interval) was not investigated (Taguchi et al., 2015). The PG0809 (Sov) gene was re-sequenced and confirmed to consist of the two combined genes PG0809 and PG0810, mis-annotated in W83 genome as separate ORFs (Saiki and Konishi, 2007). A dashed arrow denotes indirect evidence that PG0809 (Sov) and PG0811 may be co-transcribed. It was shown that sigma factor SigP (regulator of other por genes) binds to the region preceding PG0811 but not the one before PG0809 (Kadowaki et al., 2016).