Fig. 1.
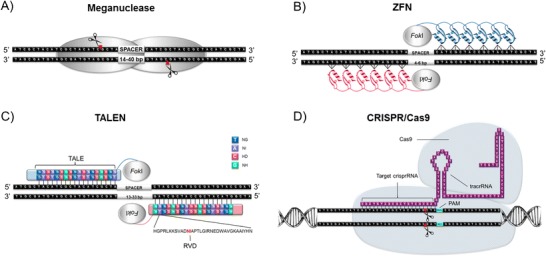
Genome editing technologies. Introduction of double-stranded breaks enables the formation of Indels in the absence of a suitable repair template, leading to knockout of gene function. Several genome editing technologies are currently available, each with a specific mode of action. a Meganucleases are homing endonucleases that form dimers in order to cleave. The single nuclease domain is made up of the DNA recognition and cleavage domains. (from Bertoni C. Front. Physiol. 2014, 5:148) [10]. b ZFNs require dimerization of two Fok1 domains at targeted loci in order for scission to occur. Each zinc finger contacts three nucleotides of the target sequence. (from Didigu CA, Doms RW. Viruses 2012, 4(2), 309–324) [11]. c TALEN cleavage is also FokI mediated; however, each TALE contains 34 amino acid repeat sequences, with each RVD targeting a single base in the target sequence. (reprinted by permission from Macmillan Publishers Ltd.: Hyongbum K, Jin-Soo K. Nature Reviews Genetics 2014, 15, 321–334) [12]. d CRISPR/Cas9 technology is RNA-guided with Cas9 mediating double-stranded cleavage of the target site. The target site is flanked by a PAM sequence, with double-stranded cleavage occurring three bases upstream from this motif (from Agrotis A, Ketteler R. Front. Genet. 2015, 6:300) [13]
