Fig. 8.
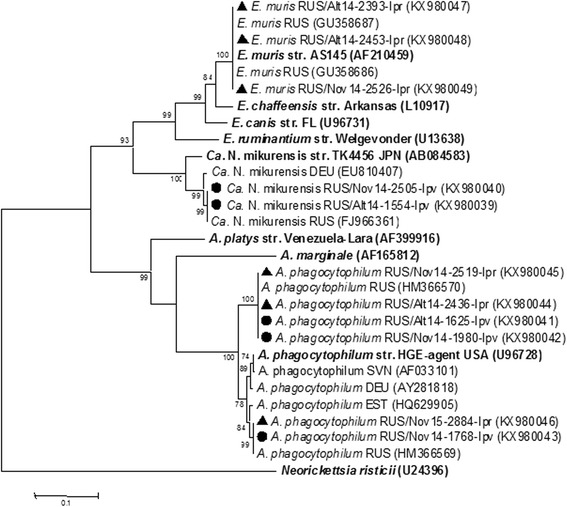
The phylogenetic tree constructed by the ML method based on nucleotide sequences of 1166–1174 bp fragment of the groESL operon of Anaplasmataceae bacteria. The scale-bar indicates an evolutionary distance of 0.1 nucleotides per position in the sequence. Significant bootstrap values (>75%) are shown on the nodes. The sequences of prototype strains of Anaplasmataceae bacteria from GenBank database are in boldface. Legend: ● I. pavlovskyi ticks; ▲ I. persulcatus ticks
