Fig. 3.
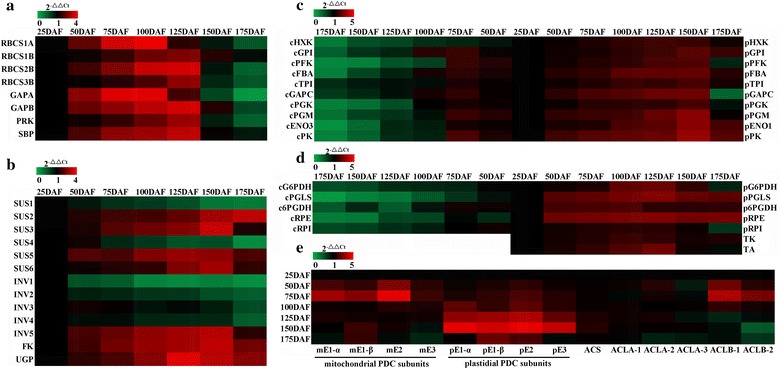
Transcriptional expression analysis for enzymes involved in carbon assimilation and partitioning in developing L. glauca fruits by qRT-PCR. a Differential expressions for genes involved in carbon assimilation. b Differential transcript patterns for enzymes related to sucrose cleavage. c Comparative analysis of transcript levels for enzymes in both cytosolic and plastidial glycolysis. d Comparative analysis of transcript levels for enzymes in both cytosolic and plastidial OPPP. e Differential transcript profiles for alternative enzymes involved in acetyl-CoA generation. The genes encoding for large subunit ribosomal protein L32e and ubiquitin-conjugating enzyme (UBC) were used as internal controls. The relative expression values in heatmap were counted as 2−△△Ct. The cytosolic (c), plastidial (p), or mitochondrial (m) isoforms of the enzymes are indicated by a prefix in c, d, or e, respectively
