Abstract
Cilia appear to be derived, evolutionarily, from structures present in the ancestral (pre-ciliary) eukaryote, such as microtubule-based vesicle trafficking and chromosome segregation systems. Experimental observations suggest that the ciliary gate, the molecular complex that mediates the selective molecular movement between cytoplasmic and ciliary compartments, shares features with nuclear pores. Our hypothesis is that this shared transport machinery is at least partially responsible for the observation that a number of ciliary and ciliogenesis-associated proteins are found within nuclei where they play roles in the regulation of gene expression, DNA repair, and nuclear import and export. Recognizing the potential for such nuclear roles is critical when considering the phenotypic effects that arise from the mutational modification of ciliary proteins.
Electronic supplementary material
The online version of this article (doi:10.1186/s13630-017-0052-x) contains supplementary material, which is available to authorized users.
Introduction
Cilia, whether immotile (primary) or motile, flagella (on sperm), or the cilia-based structures within photoreceptor cells, represent a distinct cytoplasmic and plasma membrane domain [26]. As has been reviewed recently, mutations in cilia- or ciliogenesis-associated proteins can lead to a range of human pathologies, known collectively as ciliopathies [8, 26, 31, 49, 64, 76, 86, 88, 91, 94]. In addition to their well appreciated cilia- and ciliogenesis-associated roles, it is now increasingly apparent that cilia-associated proteins have non-ciliary, cytoplasmic roles, described in a number of recent reviews [87, 93, 102]. For example, it is well established that cilia and cilia-associated proteins are involved in Hedgehog, TGF-β, Wnt, PDGFRα, Notch, and Hippo inter-cellular signaling systems [12, 31, 36, 62, 65, 86]. That said, cilia themselves may not be strictly necessary for a number of these signaling pathways. For example, myeloid and lymphoid cells reportedly lack cilia (primary or motile) yet express cilia-associated proteins [24] and support Hedgehog signaling [16, 93]. Similarly, in the zebrafish, Huang and Schier [37] found that genetic abolition of maternal and zygotic cilia formation left Wnt signaling intact while dysregulating hedgehog signaling.
In addition to their signaling functions, there is clear evidence for the role of a number of cilia-associated proteins in DNA damage repair [4, 102]. In this review, we highlight the observation that a number of cilia-associated proteins have a nuclear presence, and in some cases, functional nuclear roles. We consider the possibility that the nuclear functions of cilia-associated proteins, arising in part from their evolutionary ancestry, can contribute to ciliopathic phenotypes through both developmentally specific and more generic mechanisms.
Implications of the evolutionary origin of cilia
While speculating about processes that occurred billions of years ago can be problematic, there is a clear consensus that the evolution of cilia began in an ancestor that contained a nucleus, with nuclear pores mediating molecular movement across the nuclear envelope and microtubule-based systems involved in chromosome segregation [10, 27, 58, 59, 70]. Entry into the ciliary domain is regulated through a distinct “transition zone” between non-ciliary cytoplasmic and plasma membrane domains. The details of this ciliary gate are beginning to be resolved [17, 39]. Resendes et al. [73] reported that the basal body protein centrin-2 (Cetn2) interacts with the nucleoporin (Nup) 107–160 complex, an integral component of the nuclear pore. As with the yeast homolog Cdc31 [25, 74], Cetn2 is an integral component of the nuclear pore. The observation that nuclear transport proteins and nuclear localization-like sequences are involved in ciliary transport [19, 38, 83] and that blocking nuclear transport also blocks ciliary import [43, 82] suggests that nuclear pore/transport proteins were co-opted during ciliary evolution. That said, Breslow et al. [7] failed to find Nup localization or a functional role for Nups in the passive permeability barrier of primary (immotile) cilia. It is clear that that this ciliary permeability barrier/transport system, which we refer to as the ciliary gate, is structurally and functionally distinct from the nuclear pore complex. In their studies of various motile cilia in the developing Xenopus embryo and cultured human retina pigment epithelial (RPE) cells, del Viso et al. [17] found evidence that the association of the inner ring (of the nuclear pore complex) Nups 188 and 93, but not other Nups examined (e.g., Nup63), are required for the formation of motile cilia in a range of cell types (the authors did not examine immotile, primary cilia). Their structural studies localized Nups 188/93 to the cilia base region, while 3D super-resolution microscopy in RPE cells revealed “the presence of two discrete structures, each composed of dozens of individual localization clusters that could not be resolved by confocal microscopy.” The overall distribution of cilia-associated Nup188/93 is distinctly different from the distribution of these proteins at the nuclear pore.
In the light of these studies, there is a general possibility that a subset of cilia-associated proteins contain localization/interaction sequences that can be recognized by cilia-associated structures, such as the Nups found in the pericentriolar material (PCM) region of the ciliary base, as well as components of the nuclear pore complex. The association of RAN complex components, important in active transport through nuclear pores [61], in the transition zone of cilia supports this model [19, 21]. RAN appears to be involved in “controlling injections of IFT proteins into the flagellar compartment” and “is, therefore, crucial to ciliogenesis” [53]. Similarly, transport of Kif17 [19, 29], retinitis pigmentosa 2 protein (RP2) [38], and Gli2 [89] into cilia is reported to be importin-β2, but not importin-α/β1, dependent (in the case of Gli2).
As examples, both the ciliary axoneme-associated radial spoke protein 3 (RSPH3–OMIM: 615876) [100] and the pericentriolar and ciliary basal body-associated protein pericentrin (PCNT: OMIM: 605925) [51] have been found to contain functional nuclear localization (NLS) and nuclear exclusion (NES) sequences. Similarly parafusin, a signaling scaffolding protein, has been found localized to the base of primary cilia in a variety of mammalian cell types and within the nuclei of fibroblasts [77]. The question arises then how common is it to find cilia-associated proteins in nuclei and what roles, if any, do they play there?
Characterizing cilia-associated proteins
Eukaryotic cilia are complex organelles composed of modified (double and triplet) microtubules and various associated proteins. Ishikawa et al. [41] identified 195 polypeptides in primary cilia, of which ~75% appeared to be present in motile cilia as well. Proximity labeling studies [57] have identified over 370 cilia-associated proteins. Boldt et al. [5] used an affinity proteomic-based study to identify 1319 cilia-associated proteins, 4905 inter-polypeptide interactions, and 52 molecular complexes. Such interaction complexity highlights the heterogeneous nature of ciliary functions in specific cell types and the likelihood that multiple evolutionary events along the vertebrate lineage have led to multiple functions for many of these proteins.
It is important to consider, however, that just because a protein is found associated with other ciliary components does not mean that its primary or sole function is ciliary. Many proteins are poorly studied, apparently for largely historical rather than functional criteria; Pandey et al. [68] coined the term “ignorome” for such proteins (and the genes that encode them). A dramatic example of the difficulty in defining gene product functions (one basis for the ignorome) is illustrated by the studies of Hutchinson et al. [40] who produced a minimal bacterial genome (JCV-syn3.0) containing 473 genes. Analysis of JCV-syn3.0 revealed an ignorome of 149 genes of unknown function (~32% of the total genome); these are genes required to produce a viable organism. At the same time, there is an increasing awareness that a number of proteins, originally identified as structural or functional components of one cellular system, can have quite distinct functions in another; they are what we might term “multi-taskers.” An example of such multitasking proteins is the catenins. Originally identified in the context of cadherin-based cell adhesion junctions [67], catenins were subsequently recognized to play important roles in the extracellular signaling-mediated regulation of gene expression [47, 54]. In this light, Kustatscher and Rappsilber [48] suggested that proteins have a certain degree of “fuzzy” distribution within cells. This notion, strengthened by proteomic analyses of subcellular compartments and organelle fractionation, indicates that stochastic intracellular movements are evolutionarily advantageous, as they increase the interaction potential of multitasker proteins. Contextually advantageous interactions are eventually “captured” by positive selection. This may well be the case for ciliary proteins in the nucleus, where their shared functions within related structures put them in position to acquire novel roles under specific circumstances.
One approach to defining the extent to which cilia-associated proteins are localized to nuclei involves mass spectrometric–proteomic studies of isolated nuclei. There are often technical obstacles to overcome in such an analysis, including the possibility of protein leakage and redistribution during nuclear isolation (see [55]). In their study Wühr et al [99] exploited unique aspects of the Xenopus laevis oocyte nucleus (the germinal vesicle), specifically its large size (~400 μm in diameter) and the ability to isolate intact germinal vesicles rapidly from late stage oocytes (Fig. 1) [46]. At the same time, oocytes are not known to express cilia of any kind, which limits the analysis to those proteins expressed during later stages of oogenesis. Wühr et al. characterized proteins based on their “relative nuclear concentration” (RNC), defined as the ratio of concentrations in the nucleus to the concentrations in nucleus plus the cytoplasm“; the RNC ranges from 1 for a totally nuclear protein to 0 for a completely cytoplasmic (non-nuclear) protein. Of course polypeptides that reside in the nucleus are synthesized in the cytoplasm, nuclear isolation is not completely “clean” (even with isolated germinal vesicles), and a number of proteins are known to shuttle between nuclear and cytoplasmic compartments, so most RNC values are intermediate between 0 and 1. Nevertheless, it is possible to distinguish classes of proteins that appear to be excluded from nuclei and confined to the cytoplasm (value of RNC <0.1) from those that have a significant nuclear localization signal (RNC >0.35).
Fig. 1.
Isolated Xenopus germinal vesicles (image from [46]); each oocyte has a diameter of ~1 mm
To estimate the number of cilia-associated proteins localized to nuclei, we used the Wühr et al. dataset together with the “gold standard” set of ~300 cilia-associated polypeptides described by van Dam et al. [92]1; to this database, we added polypeptides known to be involved in ciliary function, i.e., C2orf59 [42], Cetn2 [81, 90, 101], and EFHC1 and EFHC2 (see [103]), for a total of 307 polypeptides. Of these 307 “gold standard” ciliary proteins, Wühr et al. report data on 118; of these, 30 (~25%) have RNC values of >0.35 (Table 1) and so are expected to have a significant nuclear presence. Other 29 polypeptides (~24%) have RNC values between 0.35 and 0.1 and may well be able to enter the nuclei. Within subgroups of related proteins, such as the interflagellar transport polypeptides (IFTs), most appear exclusively cytoplasmic, but a few (e.g., IFT27) (Table 1) seem to be present at significant levels within the germinal vesicle. Similarly, the radial spoke protein RSPH4A (OMIM: 612647) has an RNC value of 0.9, higher than the known transcription factor SOX13 (0.84), suggesting that RSPH4A is strongly concentrated in the germinal vesicle. In contrast, β-catenin (CTNNB1), which is well known to be able to enter the nuclei in response to canonical Wnt signaling (see [54]), appears to be exclusively cytoplasmic (RNC = 0.031) in the oocyte.
Table 1.
Ciliary proteins with a nuclear presence
| Nuclear + cytoplasmic >0.2 | Ambiguous between 0.2 and 0.1 | Cytoplasmic >0.1 |
| ARL3 | ADP-ribosylation factor-like protein 3 | RNC = 0.51 (nuc + cyto) |
| Interactor with RP2 and PDEδ. Involved in trafficking of vesicles from the Golgi to the cilium, especially; farnesylated cargo in association with PDEδ and RPGR; and myristoylated cargo in association with UNC119 and RP2 | ||
| ARL6 | ADP-ribosylation factor-like protein 6 | RNC = 0.49 (nuc + cyto) |
| Mutated in Bardet–Biedl syndrome (type 3) and retinitis pigmentosa (type 55). At the ciliary gate, it regulates Wnt signaling. Functions with BBSome to coat proteins for trafficking to the cilium basal body, cilium, cytosol, transition zone | ||
| ATXN10 | Ataxin 10 | RNC = 0.48 (nuc + cyto) |
| Mutated in 1 NPHP family (splice-site mutation). Forms complex with NPHP5 and 6 at the basal body | ||
| C21orf59 a | Jaffe et al. [42] c21orf59/kurly controls both cilia motility and polarization | RNS = 0.48 (nuc + cyto) |
| C8orf37 | Mutated in cone–rod dystrophy (type 16) and retinitis pigmentosa (type 64). Localizes to basal body in cultured RPE cells and Basal body and ciliary rootlet in mouse photoreceptors Basal body, ciliary root |
RNS = 0.36 (nuc + cyto) |
| CETN2 a | Centrin-2 | RNS = 0.55/0.58 (nuc + cyto) |
| CETN3 a | Centrin-3 | RNS = 0.33 (nuc + cyto) |
| CTNNB1 | Catenin beta-1 | RNS = 0.031 (cytoplasmic a ) |
| Involved in regulation of PKD1 and PKD2 expression. Nek2 substrate involved in centrosome separation, along with rootletin (CROCC). Facilitator of canonical Wnt signalling pathway. Many links between cilia and Wnt signaling centrosome | ||
| DNAL1 | Dynein light chain 1, axonemal | RNS = 0.49 (nuc + cyto) |
| Mutated in primary ciliary dyskinesia (type 16) Component of outer dynein arms. Axoneme | ||
| DPCD | Deleted in primary ciliary dyskinesia | RNS = 0.29 (nuc + cyto) |
| Deleted in a mouse model of primary ciliary dyskinesia | ||
| EFHC2 a | EF-hand domain-containing family member C2 | RNS = 0.47 (nuc + cyto) |
| HEATR2 | HEAT repeat-containing protein 2 | RNS = 0.63 (nuc + cyto) |
| Mutation linked to premature centromere division (PCD), presumably involved in dynein arm transport or assembly | ||
| HSPA8 | Heat shock cognate 71 kDa protein | RNS = 0.49 (nuc + cyto) |
| Chaperone of the IFT together with DnajB6 | ||
| HSPB11 | Heat shock protein beta-11 | RNS = 0.48 (nuc + cyto) |
| IFT25, part of IFT-B complex. Forms a complex with IFT27. External submission. Cilium, IFT | ||
| IFT27 | Intraflagellar transport protein 27 | RNS = 0.45 (nuc + cyto) |
| Component of IFT complex B. Rab-like small G protein basal body, cilium, IFT | ||
| KIF17 | Kinesin-like protein KIF17 | RNS = 0.21 (nuc + cyto) |
| Kinesin 2 motor, active in antergrade IFT ciliary tip | ||
| LRRC6 | Protein TILB homolog | RNS = 0.34 (nuc + cyto) |
| Essential for proper axonemal assembly of inner and outer dynein arms, causes PCD. Cilium | ||
| MAPRE1 (EB1) | MT-associated protein RP/EB family 1 | RNS = 0.38 (nuc + cyto) |
| MT plus-end-tracking protein. promotes ciliogenesis. Centrosome, golgi | ||
| MNS1 | Meiosis-specific nuclear structural protein 1 | RNS = 0.37 (nuc + cyto) |
| Mns1−/− mice have short, immotile sperm flagella, situs defects, and hydrocephalus. Protein localized along flagellum. Knockdown in IMCD3 and 3T3 cells causes Hh signaling defects axoneme | ||
| NME7 | Nucleoside diphosphate kinase 7 | RNS = 0.21 (nuc + cyto) |
| IFT transport and signaling defects after knockdown | ||
| NME8 | Thioredoxin domain-containing protein 3 | RNS = 0.39 (nuc + cyto) |
| The sea urchin ortholog of this gene encodes a component of sperm outer dynein arms, and the protein is implicated in ciliary function. Mutations in this gene are implicated in primary ciliary dyskinesia type 6 | ||
| NPHP1 | Nephrocystin-1 | RNS = 0.36 (nuc + cyto) |
| Known ciliopathy gene JBTS4, NPHP1, SLS1 transition zone | ||
| NUP214 | Nuclear pore complex protein Nup214 | RNS = 0.45 (nuc + cyto) |
| Part of ciliary pore complex transition zone | ||
| NUP35 | Nucleoporin NUP35 | RNS = 0.47 (nuc + cyto) |
| Nucleoporin 35 kDa, part of ciliary pore complex transition zone | ||
| NUP37 | Nucleoporin NUP37 | RNS = 0.56 (nuc + cyto) |
| Part of ciliary pore complex transition zone | ||
| NUP62 | Nuclear pore glycoprotein p62 | RNS = 0.38 (nuc + cyto) |
| Part of ciliary pore complex transition zone | ||
| NUP93 | Nuclear pore complex protein Nup93 | RNS − 0.45 (nuc + cyto) |
| Part of ciliary pore complex transition zone | ||
| OCRL | Oculocerebrorenal syndrome of Lowe | RNS = 0.24 (nuc + cyto) |
| Linked to Lowe Syndrome. Involved in assembly of primary cilia, involved in Rab8n-dependent protein trafficking to the cilium | ||
| ORC1 | Origin recognition complex subunit 1 | RNS = 0.23 (nuc + cyto) |
| Linked to Meier–Gorlin syndrome (MGS), a disorder conferring microcephaly, primordial dwarfism, underdeveloped ears, and skeletal abnormalities. knockdown affects ciliogenesis and Hh signaling | ||
| PAFAH1B1 | Platelet-activating factor acetylhydrolase IB alpha | RNS = 0.28 (nuc + cyto) |
| LIS1 Mammalian Lis1 localizes to motile cilia in trachea and oviduct, but is absent from non-motile primary cilia axoneme | ||
| PDE6D | cGMP 3′,5′-cyclic phosphodiesterase subunit delta | RNS = 0.40 (nuc + cyto)p |
| Part of the ARL13B, INPP5E, and CEP164 network | ||
| PLK1 | Serine/threonine-protein kinase PLK1 | RNS = 0.39 (nuc + cyto) |
| Localizes to TZ and induces phosphorylation of NPHP1 transition zone | ||
| RABL5 | Rab-like protein 5 | RNS = 0.33 (nuc + cyto) |
| IFT22, component of IFT complex B cilium, IFT | ||
| RILPL2 | RILP-like protein 2 | RNS = 0.50 (nuc + cyto) |
| Rab effector. Regulates cilium membrane content. Cilium, basal body | ||
| RSPH4A | Radial spoke head protein 4 homolog A | RNS = 0.90 (most nuclear) |
| Defective Hh signaling but no structural cilia defects after knockdown. Mutations in RSPH4A cause primary ciliary dyskinesia, with typical respiratory features, but without situs abnormalities. These findings suggest that radial spoke proteins are not essential for embryonic nodal ciliary function, which is important in the determination of left–right axis development cilium, axoneme | ||
| RSPH9 | Radial spoke head protein 9 homolog | RNS = 0.44 (nuc + cyto) |
| Mutations in RSPH9 linked to primary ciliary dyskinesia, with typical respiratory features, but without situs abnormalities. These findings suggest that radial spoke proteins are not essential for embryonic nodal ciliary function, which is important in the determination of left–right axis development structural cilia defects of primary cilia after knockdown in 3 different murine cell lines cilium, axoneme | ||
| SNX10 | Sorting nexin-10 (Fragment) | RNS = 0.51 (nuc + cyto) |
| Regulates ciliogenesis Centrosome | ||
| SSNA1 | Sjoegren syndrome nuclear autoantigen 1 | RNA + 0.50 (nuc + cyto) |
| siRNA knockdown in 3 mouse cell lines perturbs receptor transport into cilium and HH signaling | ||
| STK38L | Serine/threonine-protein kinase 38-like | RNA = 0.34 (nuc + cyto) |
| Phosphorylates Rabin8. Mutated in canine retinal degeneration. Cytosol | ||
| SUFU | Suppressor of fused homolog | RNS = 0.24 (nuc + cyto) |
| Localizes to ciliary tip together with GLI transcription factors. Role in Hh signaling ciliary tip | ||
| TEKT4 | Tektin-4 | RNS = 0.35 (nuc + cyto) |
| Axonemal protein required for flagella motility in mouse sperm. Expression reduced in inasthenozoospermic men. Cilium | ||
| TNPO1 | Transportin-1 | RNS = 0.44 (nuc + cyto) |
| Importin beta 2. Regulates entry of RP2 and kinesin motor into cilium axoneme | ||
| TRAPPC3 | Trafficking protein particle complex subunit 3 | RNS = 0.30 (nuc + cyto). |
| Required for Rabin8 centrosome trafficking and ciliogenesis basal body, centrosome | ||
| TTK | Dual specificity protein kinase TTK | RNS = 0.33 (nuc + cyto) |
| Negatively regulates ciliogenesis. Centrosome | ||
| WDR19 | WD repeat-containing protein 19 | RNS = 0.22 (nuc + cyto) |
| Mutations in WDR19 associated with ciliopathies nephronophthisis (NPHP13), Jeune and Sensenbrenner syndromes (ATD5), IFT complex A component (aka IFT144) cilium, IFT |
Xenopus Proteomic data from Wuhr et al. ciliary proteins (and curator notes) from: http://www.syscilia.org/goldstandard.shtml
aIndicates known ciliary protein added to list of ciliary proteins
In a related set of unpublished studies, “nuclear” fractions were prepared from human SH-SY5Y neuroblastoma cells and analyzed by mass spectrometry (Additional file 1); of the 307 “gold standard” ciliary proteins, 93 were found within this nuclear (Table 2) fraction, including 15 that also had a “nuclear presence” in the Xenopus germinal vesicle dataset (Additional file 1). Together these data support the hypothesis that a significant percentage of the polypeptides linked to cilia formation, function, and phenotypic defects appear to be able to enter, and potentially play functional roles, in the nucleus. At this point, we describe a few of the best characterized of these nuclear functions.
Table 2.
Cilia-associated proteins found in SHSY5Y-NUCLEAR dataset
| ARF4 | ARL3a | ARL6a | ASAP1 | ATXN10a | Cetn2a |
| Cetn3 | CEP135 | CEP250 | CEP41 | CEP89 | CEP97 |
| CP110 | CROCC | CTNNB1b | DNAH1a | DNAH11 | DNAH5 |
| DPCD | DPYSL2 | DYNLT1 | EXOC3 | EXOC4 | EXOC5 |
| EXOC6 | FLNA | GSK3B | HEATR2a | HSPA8a | HTT |
| IFT27a | IFT52 | IFT81 | INVS | KIF3A | KIF3C |
| MLF1 | NGFR | NME7 | MAPRE1a (EB1) | LZTFL1 | NPHP4 |
| NUP214a | NUP35a | NUP37 | NUP62a | NUP93a | OCRL |
| ODF2 | ORC1 | PAFAH1B1 | PARD3 | PCM1 | PDE6 Da |
| PHF17 | PKD2 | PLK1a | RAB11A | RAB23 | RAB8A |
| RAN | RANBP1 | RFX3 | RP2 | SEPT2 | SEPT7 |
| SGK196 | SNAP25 | STK38L | SYNE2 | TNPO1a | TRAPPC10 |
| TRAPPC3 | TRIM32 | TRIP11 | TTC21B | TTC30A | TTC8 |
| TUBA1C | TUBA4A | TUBB2A | TUBB2B | TUBB3 | TUBE1 |
| TUBGCP2 | TUBGCP3 | TUBGCP4 | VDAC3 | XPNPEP3 |
aIndicates also found to have a nuclear presence in the Xenopus dataset (Table 1)
bJUP/Plakoglobin also present
Examples of cilia-associated proteins with known nuclear roles
Nephronophthisis (NPH) is an autosomal recessive cystic kidney disease characterized by inflammation and scarring (fibrosis) that compromises kidney function and leads, over time, to end-stage renal disease [98]. Many of the genes linked to NPH, encoding polypeptides known as nephrocystins or NPHPs, have been found to encode proteins localized to or involved in primary cilia formation or cilia-dependent signaling (both Wnt and Shh) which presumably explains phenotypic effects on other organ systems. Where other organs are involved, the disease is termed NPH-related ciliopathy [98]. Of the NPHP-associated genes, a number have been found to have direct or indirect nuclear roles. For example, NPHP4 (nephrocystin-2: OMIM 607215) interacts with and inhibits the LATS1 kinase, leading to the nuclear accumulation of the transcriptional co-activators YAP and TAZ, two proteins involved in the HIPPO (hypoxia) signaling system [33]. NPHP9 (also known as NEK8–OMIM 609799) has been found to interact with TAZ, leading to the nuclear localization of the NPHP9–TAXZ complex [34]. TAZ is normally exported to the cytoplasm through interactions with 14-3-3 proteins; NPHP9 appears to compete with 14-3-3 proteins for TAZ binding. NPHP4 inhibits LATS1 phosphorylation of TAZ, which reduces its affinity for 14-3-3 proteins. NPHP4 also interacts with the ubiquitin ligase JADE1 (another ciliary and nuclear component), an interaction that appears to stabilize JADE1 and lead to its nuclear localization where it acts to destabilize β-catenin and inhibit canonical Wnt signaling [6]. NPHP4 itself appears to be primarily perinuclear. The absence of NPHP4 activity leads to an increase in Wnt signaling and subsequent cyst formation in the kidney [6]. Studies in the zebrafish support this mechanism [9]. NPHP7 (OMIM: 611498) is a GLI-like zinc finger transcription factor involved in the regulation of mesenchymal–epithelial cell behavior, inhibiting the expression of genes such as Snail1 and Wnt4 [72]. Mutations in NPHP7 have been associated with NPH-related ciliopathy [35]. Morpholino-mediated downregulation of NPHP7 has been found to influence the formation of immotile motile cilia and a number of associated phenotypes, including cystic pronephros and ciliary motility in zebrafish [45, 71], presumably due to effects on gene expression. NPHP10 (OMIM: 613524) localizes to nuclear foci with a number of DNA damage response proteins, including the centrosomal protein CEP164 (OMIM: 614848) [11].
Oral-facial-digital 1 (OFD1) syndrome
OFD1 (OMIM: 311200) is one of an array of nine phenotypically similar disorders. OFD1 is an X-linked dominant disease associated with facial malformations as well as cystic kidneys [22]. OFD1 has been linked to the gene OFD1 (previously known as Cxorf5) (OMIM: 300170). In a mouse model OFD1 has been associated with defects in primary cilia formation and left–right axis formation [23]. Subsequent studies indicate that OFD1 has a nuclear presence, and plays a role in chromatin remodeling [32] and in double-stranded DNA break repair, mediated by the TIP60 complex [1].
Bardet-Biedl Syndrome
Bardet–Biedl syndromes are group of pleiotropic oligogenic ciliopathies [64] that have been linked to (at least) nineteen different genes with potentially etiological roles. Of these, BBS1, 2, 4, 5, 6, 7, 8, and 10 have been found to enter the nucleus where they can influence gene expression through interactions with the polycomb group member protein RNF2 (Ring Finger Protein 4: OMIM: 602850) [30]. Both BBS1 (OMIM: 209901) and BBS11 (OMIM: 615988) have been reported to interact with and to alter the activity of NPHP7 [45, 71].
Centrins and pericentrin
Centrins (Cetn) are calmodulin-like proteins associated with centrosomes (microtubule-organizing centers) and the basal body regions of cilia. Two centrin subclasses have been identified, Cetn2-like and Cetn3-like [14, 28]. In the yeast Saccharomyces cerevisiae, there is a single centrin gene (Cdc31); the Cdc31 protein is found localized to nuclear pores in addition to the spindle pole body [74]. Mutations in Cdc31 lead to defects in mRNA export [25]. In vertebrate cells, Cetn2 (OMIM: 300006) has been found associated with nucleoporins and localized to nuclear pores; expression of the Cetn2-binding regions of NUP160 led to a decrease in nuclear export of mRNA and proteins, without obvious effects on protein import [73].
While Cetns have been reported to play a role in cell division in vertebrate cells [75], subsequent studies in chick DT40 cells [15] revealed that null mutations in all three centrin genes had no effect on cell division (although effects on cilia formation were not reported). This made it possible to examine the nuclear functions of centrins, two of which had been previously reported. First, Cetn2 is an integral component of the nucleotide excision repair/xeroderma pigmentosum group C (XPC–RAD23–CETN2) complex [3, 63]. Cetn2-null DT40 cells displayed no defects in centrosome formation or cell division but were reported to be hypersensitive to UV irradiation [15]. The nuclear excision repair protein Rad33 appears functionally homologous to Cetn2 [18]. The centrin-associated protein pericentrin (PCNT) (OMIM: 605925) has also been implicated in DNA damage repair [96]. Interactions between pericentrin and microcephalin (MCPH1) have been implicated in primordial dwarfism [60], genome instability, and centrosome amplification [2].
More recently, Cetn2 has been found to be part of the nuclear pore-associated TREX-2 complex (GANP, DSS1, ENY2, PCID2, & CETN2) [13]. TREX-2, in turn, has been reported to interact with the Mediator complex, linking the regulation of gene expression and the export of mRNAs from the nucleus [78, 79]. Our own work in Xenopus indicates that Cetn2 associates with sites within the promoter regions of a subset of FGF and FGF receptor genes, regulating their expression and that its morpholino-mediated downregulation leads to defects in mesoderm formation [81].
An interesting point to emerge from studies of Cetn2 is that while standard immunofluorescence microscopy reveals it to be concentrated at centrosomes and basal bodies, cell fractionation studies suggest that more than 90% of centrin is soluble [69] and associated with the xeroderma pigmentous complex (XPC) nucleotide excision repair complex [3, 63], a reminder that proteins can redistribute, often dramatically, upon cell solubilization (see [55]).
Chibby (Cby1) (OMIM: 607757) is a small evolutionarily conserved protein associated with basal bodies and involved in ciliogenesis [20]. In humans, there are three Cby-like proteins (Cby, Cby2, and Cby3). In vertebrate cells but not (apparently) in Drosophila [20], Cby acts as a negative regulator of β-catenin-mediated Wnt signaling [80, 84, 85].2 Cby’s interaction with β-catenin involves 14-3-3 proteins and leads to β-catenin’s export from the nucleus [44, 50]. STRING analysis (Fig. 2) indicates that Cby1 interacts with a number of 14-3-3 (YWHAX) proteins; a conclusion supported by preliminary studies in which Cby-GFP was found to co-precipitate with a number of YWHAX proteins in Xenopus (stars in Fig. 2) (McClure-Begley et al. unpublished observations). In human-induced pluripotent stem cell (HiPSC)-derived cerebral organoids, Cby is widely expressed and appears largely nuclear (McClure-Begley et al. in progress) (Fig. 3). Characterization of cby −/− mice (in a C57BL/6 background) showed that ~75% died within 2 weeks of birth and were “runted and demonstrated anemia” [95]; those animals that survived displayed a number of cilia-related defects [52]. In X. laevis, morpholino-mediated downregulation of Cby led to ciliary defects and abnormal neural crest, central nervous system, and pronephros development [80]. Many, but not all, morphant phenotypes could be ameliorated by the extracellular Wnt inhibitor SFRP2, suggesting that these Cby deficit phenotypes are due to increased Wnt signaling. In this light, Cby morphant effects on Hh components were not rescued by SFRP2, and so presumably represent other (that is, non-canonical Wnt signaling related) processes [80]. A similar behavior appears to be displayed by the 92 kDa nuclear form of Inversin (OMIM: 243305), which has been found associated with β-catenin [66]. Whether, like other β-catenin-interacting proteins [54, 97], Cby1 and Inversin also interact with the paralogous protein plakoglobin (ɣ-catenin or JUP: OMIM: 173325) has not been reported.
Fig. 2.
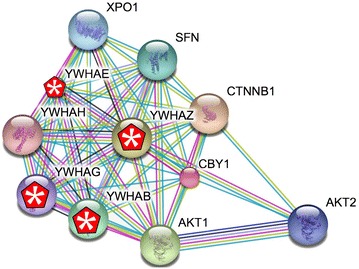
A STRING interaction map for Cby1 (http://string-db.org/cgi/network.pl?taskId=nnIlPEaytPOX) reveals a number of interaction partners. 14-3-3 proteins (YHYAX) marked by * were identified as Cby1-associated proteins in Xenopus laevis using an immunoisolation and mass spectrometry analysis (Shi et al. unpublished observations)
Fig. 3.
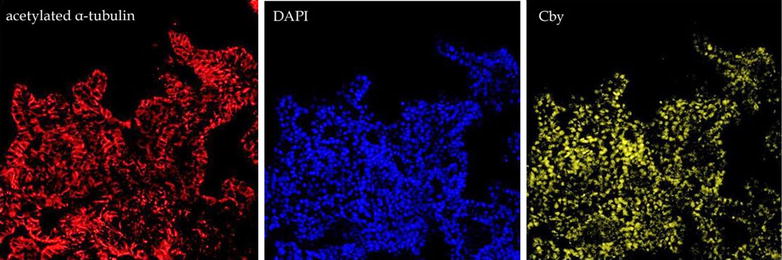
Human iPSC-derived cerebral organoids were stained in whole-mount for acetylated α-tubulin (lefthand panel), the DNA marker DAPI (central panel), and Cby (righthand panel) using a rabbit anti-Cby antibody, supplied by Feng-Qian Li (University of Stonybrook) and described in Li et al. [50]
Summary
The challenges in separating the ciliary and cytoplasmic from possible nuclear functions of cilia-associated proteins are similar to those faced in the analysis of the adhesive (cytoplasmic) and gene expression (nuclear) roles of β-catenin and γ-catenin (plakoglobin), which share many interaction partners [97]. The use of cytoplasmically anchored forms allowed us to conclude that γ-catenin’s effects on β-catenin-mediated gene expression were indirect [56]. Given the shared mechanisms acting at nuclear pores and ciliary gates, a similar (or perhaps a more modern) strategy seems necessary to distinguish the ciliary and cytoplasmic from the non-ciliary, i.e., nuclear, functional roles of cilia-associated proteins.
Authors’ contributions
The manuscript was conceived and written collaboratively by the two authors. Both authors read and approved the final manuscript.
Acknowledgements
We thank William Old, Chris C. Ebmeier, Douglas A. Chapnick, and Kerri E. Ball for sharing their data on the proteomic analysis of nuclear fractions of SH-SY5Y neuroblastoma cells (Additional file 1). We thank Jianli Shi, Ying Zhao, and Mark Winey for support and discussion over the past few years.
Competing interests
Both authors declare that they have no competing interests.
Availability of data
Results from the proteomic analysis of SH-SY5Y cells will be made publicly available through the Old Lab web server.
Funding
Previous work on cilia in Xenopus was supported in part by a supplement to NIH Grant GM074746. Proteomic analysis of SH-SY5Y cells was carried out under a cooperative agreement with DARPA (13-34-RTA-FP-007) to William Old. Studies on human iPSC-derived organoids have been supported by grants from the Linda Crnic Foundation to Tristan McClure-Begley, William Old, and Michael Klymkowsky.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- OMIM
Online Mendelian Inheritance in Man: https://www.omim.org
- RNC
relative nuclear concentration
- RPE cells
human retina pigment epithelial cells
- NLS
nuclear localization sequence
- NES
nuclear exclusion sequence
Additional file
Additional file 1. Additional material.
Footnotes
Website accessible here: http://www.syscilia.org/goldstandard.shtml.
Based on a simple BLAST search, there does not appear to a Caenorhabditis elegans ortholog of Chibby.
Electronic supplementary material
The online version of this article (doi:10.1186/s13630-017-0052-x) contains supplementary material, which is available to authorized users.
Contributor Information
Tristan D. McClure-Begley, Email: tristan.mcclure-begley@colorado.edu
Michael W. Klymkowsky, Email: michael.klymkowsky@colorado.edu
References
- 1.Abramowicz I, Carpenter G, Alfieri M, Colnaghi R, Outwin E, Parent P, Thauvin-Robinet C, Iaconis D, Franco B, O’Driscoll M. Oral-facial-digital syndrome type I cells exhibit impaired DNA repair; unanticipated consequences of defective OFD1 outside of the cilia network. Hum Mol Genet. 2016: ddw364. [DOI] [PubMed]
- 2.Antonczak A, Mullee L, Wang Y, Comartin D, Inoue T, Pelletier L, Morrison C. Opposing effects of pericentrin and microcephalin on the pericentriolar material regulate CHK1 activation in the DNA damage response. Oncogene. 2016;35:2003–2010. doi: 10.1038/onc.2015.257. [DOI] [PubMed] [Google Scholar]
- 3.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 4.Attanasio M. Ciliopathies and DNA damage: an emerging nexus. Curr Opin Nephrol Hypertens. 2015;24:366–370. doi: 10.1097/MNH.0000000000000134. [DOI] [PubMed] [Google Scholar]
- 5.Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TMT, Texier Y, van Beersum SE, Horn N, Willer JR, Mans DA. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun. 2016;7:11491. doi: 10.1038/ncomms11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgal L, Habbig S, Hatzold J, Liebau MC, Dafinger C, Sacarea I, Hammerschmidt M, Benzing T, Schermer B. The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate β-catenin signaling. J Biol Chem. 2012;287:25370–25380. doi: 10.1074/jbc.M112.385658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J, Witman G. Cilia and diseases. Bioscience. 2014;64:1126–1137. doi: 10.1093/biosci/biu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burcklé C, Gaudé H-M, Vesque C, Silbermann F, Salomon R, Jeanpierre C, Antignac C, Saunier S, Schneider-Maunoury S. Control of the Wnt pathways by nephrocystin-4 is required for morphogenesis of the zebrafish pronephros. Hum Mol Genet. 2011;20:2611–2627. doi: 10.1093/hmg/ddr164. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, Wang H, Hurd TW, Zhou W, Cluckey A. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150:533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen ST, Pedersen LB, Schneider L, Satir P. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 2007;8:97–109. doi: 10.1111/j.1600-0854.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham CN, Schmidt CA, Schramm NJ, Gaylord MR, Resendes KK. Human TREX2 components PCID2 and centrin 2, but not ENY2, have distinct functions in protein export and co-localize to the centrosome. Exp Cell Res. 2014;320:209–218. doi: 10.1016/j.yexcr.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Dantas TJ, Daly OM, Morrison CG. Such small hands: the roles of centrins/caltractins in the centriole and in genome maintenance. Cell Mol Life Sci. 2012;69:2979–2997. doi: 10.1007/s00018-012-0961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dantas TJ, Wang Y, Lalor P, Dockery P, Morrison CG. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J Cell Biol. 2011;193:307–318. doi: 10.1083/jcb.201012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Roche M, Ritter AT, Angus KL, Dinsmore C, Earnshaw CH, Reiter JF, Griffiths GM. Hedgehog signaling controls T cell killing at the immunological synapse. Science. 2013;342:1247–1250. doi: 10.1126/science.1244689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, Reza N, Bewersdorf J, Lusk CP, Khokha MK. Congenital heart disease genetics uncovers context-dependent organization and function of nucleoporins at cilia. Dev Cell. 2016;38:478–492. doi: 10.1016/j.devcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.den Dulk B, van Eijk P, de Ruijter M, Brandsma JA, Brouwer J. The NER protein Rad33 shows functional homology to human Centrin2 and is involved in modification of Rad4. DNA Repair. 2008;7:858–868. doi: 10.1016/j.dnarep.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN-T, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enjolras C, Thomas J, Chhin B, Cortier E, Duteyrat JL, Soulavie F, Kernan MJ, Laurencon A, Durand B. Drosophila chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J Cell Biol. 2012;197:313–325. doi: 10.1083/jcb.201109148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan S, Margolis B. The Ran importin system in cilia trafficking. Organogenesis. 2011;7:147–153. doi: 10.4161/org.7.3.17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrante MI, Feather SA, Bulfone A, Wright V, Ghiani M, Selicorni A, Gammaro L, Scolari F, Woolf AS, Sylvie O. Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet. 2001;68:569–576. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, Dollé P, Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- 24.Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, Rosenbaum JL, Baldari CT. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332–1339. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer T, Rodríguez-Navarro S, Pereira G, Rácz A, Schiebel E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 26.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 27.Forterre P. The common ancestor of archaea and eukarya was not an archaeon. Archaea. 2013;2013:372396. doi: 10.1155/2013/372396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedberg F. Centrin isoforms in mammals. Relation to calmodulin. Mol Biol Rep. 2006;33:243–252. doi: 10.1007/s11033-006-9004-z. [DOI] [PubMed] [Google Scholar]
- 29.Funabashi T, Katoh Y, Michisaka S, Terada M, Sugawa M, Nakayama K. Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46–IFT56 as well as on its nuclear localization signal. Mol Biol Cell. 2017;28:624–633. doi: 10.1091/mbc.E16-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gascue C, Tan PL, Cardenas-Rodriguez M, Libisch G, Fernandez-Calero T, Liu YP, Astrada S, Robello C, Naya H, Katsanis N. Direct role of Bardet–Biedl syndrome proteins in transcriptional regulation. J Cell Sci. 2012;125:362–375. doi: 10.1242/jcs.089375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerhardt C, Leu T, Lier JM, Rüther U. The cilia-regulated proteasome and its role in the development of ciliopathies and cancer. Cilia. 2016;5:14. doi: 10.1186/s13630-016-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giorgio G, Alfieri M, Prattichizzo C, Zullo A, Cairo S, Franco B. Functional characterization of the OFD1 protein reveals a nuclear localization and physical interaction with subunits of a chromatin remodeling complex. Mol Biol Cell. 2007;18:4397–4404. doi: 10.1091/mbc.E07-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habbig S, Bartram MP, Müller RU, Schwarz R, Andriopoulos N, Chen S, Sägmüller JG, Hoehne M, Burst V, Liebau MC. NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J Cell Biol. 2011;193:633–642. doi: 10.1083/jcb.201009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habbig S, Bartram MP, Sägmüller JG, Griessmann A, Franke M, Müller RU, Schwarz R, Hoehne M, Bergmann C, Tessmer C. The ciliopathy disease protein NPHP9 promotes nuclear delivery and activation of the oncogenic transcriptional regulator TAZ. Hum Mol Genet. 2012;21:5528–5538. doi: 10.1093/hmg/dds408. [DOI] [PubMed] [Google Scholar]
- 35.Halbritter J, Porath JD, Diaz KA, Braun DA, Kohl S, Chaki M, Allen SJ, Soliman NA, Hildebrandt F, Otto EA. Identification of 99 novel mutations in a worldwide cohort of 1056 patients with a nephronophthisis-related ciliopathy. Hum Genet. 2013;132:865–884. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilgendorf KI, Johnson CT, Jackson PK. The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr Opin Cell Biol. 2016;39:84–92. doi: 10.1016/j.ceb.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang P, Schier AF. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–3098. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurd TW, Fan S, Margolis BL. Localization of retinitis pigmentosa 2 to cilia is regulated by Importin β2. J Cell Sci. 2011;124:718–726. doi: 10.1242/jcs.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurt E, Beck M. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr Opin Cell Biol. 2015;34:31–38. doi: 10.1016/j.ceb.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Hutchison CA, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa H, Thompson J, Yates JR, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaffe KM, Grimes DT, Schottenfeld-Roames J, Werner ME, Ku T-SJ, Kim SK, Pelliccia JL, Morante NF, Mitchell BJ, Burdine RD. c21orf59/kurly controls both cilia motility and polarization. Cell Rep. 2016;14:1841–1849. doi: 10.1016/j.celrep.2016.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kee HL, Dishinger JF, Blasius TL, Liu C-J, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Killoran RC, Fan J, Yang D, Shilton BH, Choy WY. Structural analysis of the 14-3-3ζ/Chibby interaction involved in Wnt/β-catenin signaling. PLoS ONE. 2015;10:e0123934. doi: 10.1371/journal.pone.0123934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YH, Epting D, Slanchev K, Engel C, Walz G, Kramer-Zucker A. A complex of BBS1 and NPHP7 is required for cilia motility in zebrafish. PLoS ONE. 2013;8:e72549. doi: 10.1371/journal.pone.0072549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klymkowsky MW, Maynell LA. MPF-induced breakdown of cytokeratin filament organization in the maturing Xenopus oocyte depends upon the translation of maternal mRNAs. Dev Biol. 1989;134:479–485. doi: 10.1016/0012-1606(89)90121-8. [DOI] [PubMed] [Google Scholar]
- 47.Klymkowsky MW, Parr B. A glimpse into the body language of cells: the intimate connection between cell adhesion and gene expression. Cell. 1995;83:5–8. doi: 10.1016/0092-8674(95)90226-0. [DOI] [PubMed] [Google Scholar]
- 48.Kustatscher G, Rappsilber J. Compositional dynamics: defining the fuzzy cell. Trends Cell Biol. 2016;26:800–803. doi: 10.1016/j.tcb.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Gleeson JG. The role of primary cilia in neuronal function. Neurobiol Dis. 2010;38:167–172. doi: 10.1016/j.nbd.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li FQ, Mofunanya A, Fischer V, Hall J, Takemaru K. Nuclear-cytoplasmic shuttling of Chibby controls beta-catenin signaling. Mol Biol Cell. 2010;21:311–322. doi: 10.1091/mbc.E09-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Q, Yu J, Zhuo X, Jiang Q, Zhang C. Pericentrin contains five NESs and an NLS essential for its nucleocytoplasmic trafficking during the cell cycle. Cell Res. 2010;20:948–962. doi: 10.1038/cr.2010.89. [DOI] [PubMed] [Google Scholar]
- 52.Love D, Li FQ, Burke MC, Cyge B, Ohmitsu M, Cabello J, Larson JE, Brody SL, Cohen JC, Takemaru K. Altered lung morphogenesis, epithelial cell differentiation and mechanics in mice deficient in the Wnt/beta-catenin antagonist Chibby. PLoS ONE. 2010;5:e13600. doi: 10.1371/journal.pone.0013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ludington WB, Wemmer KA, Lechtreck KF, Witman GB, Marshall WF. Avalanche-like behavior in ciliary import. Proc Natl Acad Sci. 2013;110:3925–3930. doi: 10.1073/pnas.1217354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCrea PD, Gottardi CJ. Beyond [beta]-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016;17:55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melan MA, Sluder G. Redistribution and differential extraction of soluble proteins in permeabilized cultured cells: implications for immunofluorescence microscopy. J Cell Sci. 1992;101:731–743. doi: 10.1242/jcs.101.4.731. [DOI] [PubMed] [Google Scholar]
- 56.Merriam JM, Rubenstein AB, Klymkowsky MW. Cytoplasmically anchored plakoglobin induces a WNT-like phenotype in Xenopus. Dev Biol. 1997;185:67–81. doi: 10.1006/dbio.1997.8550. [DOI] [PubMed] [Google Scholar]
- 57.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV. Proteomics of primary cilia by proximity labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell DR. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. In: Eukaryotic membranes and cytoskeleton. New York: Springer; 2007. p. 130–140. [DOI] [PMC free article] [PubMed]
- 59.Mitchell DR. Evolution of cilia. Cold Spring Harbor Perspect Biol. 2017;9:a028290. doi: 10.1101/cshperspect.a028290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyoshi K, Kasahara K, Miyazaki I, Shimizu S, Taniguchi M, Matsuzaki S, Tohyama M, Asanuma M. Pericentrin, a centrosomal protein related to microcephalic primordial dwarfism, is required for olfactory cilia assembly in mice. FASEB J. 2009;23:3289–3297. doi: 10.1096/fj.08-124420. [DOI] [PubMed] [Google Scholar]
- 61.Mor A, White MA, Fontoura BM. Nuclear trafficking in health and disease. Curr Opin Cell Biol. 2014;28:28–35. doi: 10.1016/j.ceb.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mourão A, Christensen ST, Lorentzen E. The intraflagellar transport machinery in ciliary signaling. Curr Opin Struct Biol. 2016;41:98–108. doi: 10.1016/j.sbi.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, Sugasawa K, Hanaoka F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol Cell Biol. 2005;25:5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novas R, Cardenas-Rodriguez M, Irigoín F, Badano JL. Bardet–Biedl syndrome: is it only cilia dysfunction? FEBS Lett. 2015;589:3479–3491. doi: 10.1016/j.febslet.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 65.Nozawa YI, Lin C, Chuang P-T. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev. 2013;23:429–437. doi: 10.1016/j.gde.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nürnberger J, Bacallao RL, Phillips CL. Inversin forms a complex with catenins and N-cadherin in polarized epithelial cells. Mol Biol Cell. 2002;13:3096–3106. doi: 10.1091/mbc.E02-04-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pandey AK, Lu L, Wang X, Homayouni R, Williams RW. Functionally enigmatic genes: a case study of the brain ignorome. PLoS ONE. 2014;9:e88889. doi: 10.1371/journal.pone.0088889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- 70.Pickett-Heaps J. The evolution of mitosis and the eukaryotic condition. Biosystems. 1974;6:37–48. doi: 10.1016/0303-2647(74)90009-4. [DOI] [PubMed] [Google Scholar]
- 71.Ramachandran H, Schäfer T, Kim Y, Herfurth K, Hoff S, Lienkamp SS, Kramer-Zucker A, Walz G. Interaction with the Bardet–Biedl gene product TRIM32/BBS11 modifies the half-life and localization of Glis2/NPHP7. J Biol Chem. 2014;289:8390–8401. doi: 10.1074/jbc.M113.534024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramachandran H, Yakulov TA, Engel C, Müller B, Walz G. The C175R mutation alters nuclear localization and transcriptional activity of the nephronophthisis NPHP7 gene product. Eur J Hum Genet. 2015;24:774–778. doi: 10.1038/ejhg.2015.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Resendes KK, Rasala BA, Forbes DJ. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol Cell Biol. 2008;28:1755–1769. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–652. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–1292. doi: 10.1016/S0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 76.Sánchez I, Dynlacht BD. Cilium assembly and disassembly. Nat Cell Biol. 2016;18:711–717. doi: 10.1038/ncb3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satir BH, Wyroba E, Liu L, Lethan M, Satir P, Christensen ST. Evolutionary implications of localization of the signaling scaffold protein Parafusin to both cilia and the nucleus. Cell Biol Int. 2015;39:136–145. doi: 10.1002/cbin.10337. [DOI] [PubMed] [Google Scholar]
- 78.Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Köhler A. The nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell. 2015;162:1016–1028. doi: 10.1016/j.cell.2015.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schubert T, Köhler A. Mediator and TREX-2: emerging links between transcription initiation and mRNA export. Nucleus. 2016;162:1–6. doi: 10.1080/19491034.2016.1169352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi J, Zhao Y, Galati N, Winey M, Klymkowsky MW. Chibby functions in Xenopus ciliary assembly, embryonic development, and the regulation of gene expression. Dev Biol. 2014;395:287–298. doi: 10.1016/j.ydbio.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi J, Zhou Y, Vonderfecht T, Winey M, Klymkowsky MW. Centrin-2 (Cetn2) mediated regulation of FGF/FGFR gene expression in Xenopus. Sci Rep. 2015;5:10283. doi: 10.1038/srep10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takao D, Dishinger JF, Kee HL, Pinskey JM, Allen BL, Verhey KJ. An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Curr Biol. 2014;24:2288–2294. doi: 10.1016/j.cub.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takao D, Verhey KJ. Gated entry into the ciliary compartment. Cell Mol Life Sci. 2016;73:119–127. doi: 10.1007/s00018-015-2058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- 85.Takemaru K-I, Fischer V, Li F-Q. Fine-tuning of nuclear beta-catenin by Chibby and 14-3-3. Cell Cycle. 2009;8:210–213. doi: 10.4161/cc.8.2.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taschner M, Lorentzen E. The intraflagellar transport machinery. Cold Spring Harbor Perspect Biol. 2016: a028092. [DOI] [PMC free article] [PubMed]
- 87.Taulet N, Delaval B. Non-ciliary functions of cilia proteins. Med Sci. 2014;30:1040–1046. doi: 10.1051/medsci/20143011019. [DOI] [PubMed] [Google Scholar]
- 88.Toriyama M, Lee C, Taylor SP, Duran I, Cohn DH, Bruel AL, Tabler JM, Drew K, Kelly MR, Kim S. The ciliopathy-associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat Genet. 2016;48:648–656. doi: 10.1038/ng.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Torrado B, Graña M, Badano JL, Irigoín F. Ciliary entry of the Hedgehog transcriptional activator Gli2 is mediated by the nuclear import machinery but differs from nuclear transport in being Imp-α/β1-independent. PLoS ONE. 2016;11:e0162033. doi: 10.1371/journal.pone.0162033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trojan P, Krauss N, Choe HW, Giessl A, Pulvermuller A, Wolfrum U. Centrins in retinal photoreceptor cells: regulators in the connecting cilium. Prog Retin Eye Res. 2008;27:237–259. doi: 10.1016/j.preteyeres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 91.Valente EM, Rosti RO, Gibbs E, Gleeson JG. Primary cilia in neurodevelopmental disorders. Nat Rev Neurol. 2014;10:27–36. doi: 10.1038/nrneurol.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Dam TJ, Wheway G, Slaats GG, Huynen MA, Giles RH. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013;2:7. doi: 10.1186/2046-2530-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vertii A, Bright A, Delaval B, Hehnly H, Doxsey S. New frontiers: discovering cilia-independent functions of cilia proteins. EMBO Rep. 2015;16:1275–1287. doi: 10.15252/embr.201540632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vertii A, Hung H-F, Hehnly H, Doxsey S. Human basal body basics. Cilia. 2016;5:13. doi: 10.1186/s13630-016-0030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Voronina VA, Takemaru K, Treuting P, Love D, Grubb BR, Hajjar AM, Adams A, Li FQ, Moon RT. Inactivation of Chibby affects function of motile airway cilia. J Cell Biol. 2009;185:225–233. doi: 10.1083/jcb.200809144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Dantas TJ, Lalor P, Dockery P, Morrison CG. Promoter hijack reveals pericentrin functions in mitosis and the DNA damage response. Cell Cycle. 2013;12:635–646. doi: 10.4161/cc.23516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Williams BO, Barish GD, Klymkowsky MW, Varmus HE. A comparative evaluation of beta-catenin and plakoglobin signaling activity. Oncogene. 2000;19:5720–5728. doi: 10.1038/sj.onc.1203921. [DOI] [PubMed] [Google Scholar]
- 98.Wolf M. Nephronophthisis and related syndromes. Curr Opin Pediatr. 2015;27:201–211. doi: 10.1097/MOP.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wühr M, Güttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ. The nuclear proteome of a vertebrate. Curr Biol. 2015;25:2663–2671. doi: 10.1016/j.cub.2015.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yan R, Hu X, Zhang W, Song L, Wang J, Yin Y, Chen S, Zhao S. The mouse radial spoke protein 3 is a nucleocytoplasmic shuttling protein that promotes neurogenesis. Histochem Cell Biol. 2015;144:309–319. doi: 10.1007/s00418-015-1338-y. [DOI] [PubMed] [Google Scholar]
- 101.Ying G, Avasthi P, Irwin M, Gerstner CD, Frederick JM, Lucero MT, Baehr W. Centrin 2 is required for mouse olfactory ciliary trafficking and development of ependymal cilia planar polarity. J Neurosci. 2014;34:6377–6388. doi: 10.1523/JNEUROSCI.0067-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuan S, Sun Z. Expanding horizons: ciliary proteins reach beyond cilia. Annu Rev Genet. 2013;47:353–376. doi: 10.1146/annurev-genet-111212-133243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou Y, Shi J, Winey M, Klymkowsky MW. Indentifying domains of EFHC1 involved in ciliary localization, ciliogenesis, and the regualtion of Wnt signaling. Dev Biol. 2016;411:257–265. doi: 10.1016/j.ydbio.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


