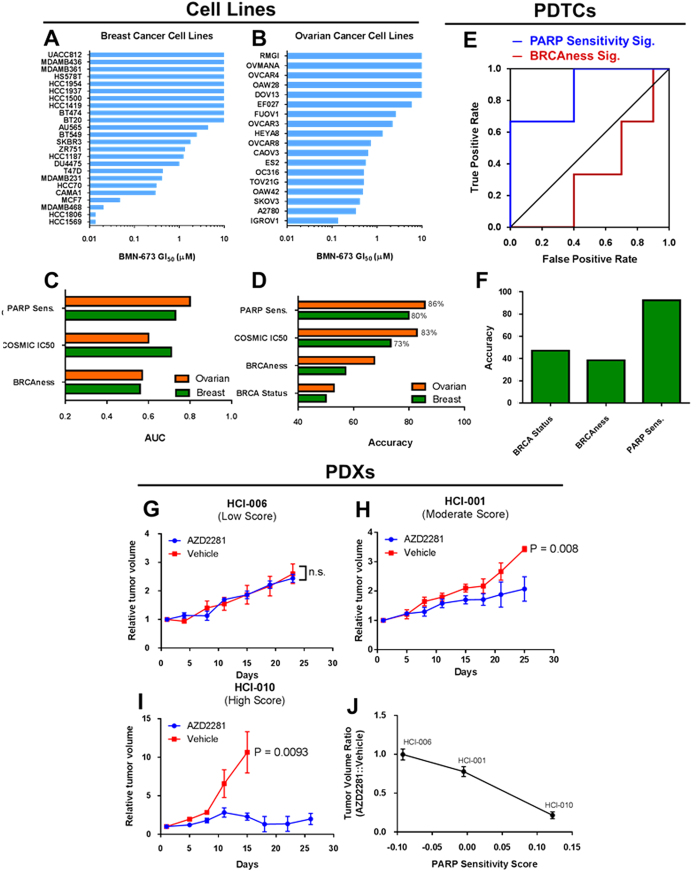
Fig. 3.
PARP sensitivity signature predicts response to PARP inhibitors in cell lines, patient-derived tumor cells (PDTCs), and patient-derived xenografts (PDXs). a, b Screening results for sensitivity to PARP inhibitor BMN-673 in breast (a) and ovarian (b) cancer cell lines. c, d AUC values from ROC curves (c) and overall accuracy (d) for prediction of sensitivity to the PARP inhibitor BMN-673 in ovarian and breast cancer cell lines determined based on BRCAness score and PARP sensitivity score, as well as by directly using the COSMIC IC50 values for AZD2281 (olaparib) and AG014699 (rucaparib) from which the PARP sensitivity signature was derived as an achievable upper bound. Overall accuracy was also analyzed based on BRCA1/2 mutation status. e ROC curves for prediction of primary PDTCs29 response to BMN-673 shows PARPi sensitivity signature outperforms BRCAness signature. f Accuracy of predicting PARPi sensitivity for PTDCs based on PARPi sensitivity score, BRCAness score, and BRCA1/2 mutation status. g–i Growth curves for breast cancer PDXs with low (g), moderate (h), and high (i) PARPi sensitivity scores following treatment with the PARP inhibitor AZD2281 QD at 50 mg/kg. j Ratio of tumor volumes in AZD2281-treated vs. vehicle controls. Ratios were calculated on day 15 and plotted as a function of the PARPi sensitivity score, demonstrating a strong negative correlation