Abstract
Plant B-box domain proteins (BBX) mediate many light-influenced developmental processes including seedling photomorphogenesis, seed germination, shade avoidance and photoperiodic regulation of flowering. Despite the wide range of potential functions, the current knowledge regarding BBX proteins in major crop plants is scarce. In this study, we identify and characterize the StBBX gene family in potato, which is composed of 30 members, with regard to structural properties and expression profiles under diurnal cycle, etiolation and de-etiolations. Based on domain organization and phylogenetic relationships, StBBX genes have been classified into five groups. Using real-time quantitative PCR, we found that expression of most of them oscillates following a 24-h rhythm; however, large differences in expression profiles were observed between the genes regarding amplitude and position of the maximal and minimal expression levels in the day/night cycle. On the basis of the time-of-day/time-of-night, we distinguished three expression groups specifically expressed during the light and two during the dark phase. In addition, we showed that the expression of several StBBX genes is under the control of the circadian clock and that some others are specifically associated with the etiolation and de-etiolation conditions. Thus, we concluded that StBBX proteins are likely key players involved in the complex diurnal and circadian networks regulating plant development as a function of light conditions and day duration.
Introduction
Plant growth and development are regulated by a wide range of environmental and intrinsic stimuli. The main external stimuli are light and temperature, whereas the fundamental internal stimuli are the circadian clock [1–3], phytohormones [4] and other growth regulatory factors [5]. Plants sense the various spectra of ambient light and convert them into biological signals through the several types of light receptors. Consecutively, photo-activated receptors modulate the activity of downstream signaling components like transcriptional regulators of light- and dark-specific genes. Such signaling pathways finally determine the timing of most developmental transitions during the plant life cycle including germination, the vegetative phase change and the floral transition [6]. On the other hand, the circadian clock synchronizes the physiological and molecular processes to the day/night cycle [1–2].
Zinc finger transcription factors comprise one of the most important family in plants. They play prominent roles in the regulation of plant growth and development. On the basis of structural and functional characteristics of individual members, they are arranged into several distinct subfamilies. Among these, B-box proteins, recently termed BBX [7], have been shown to play essential roles in the light regulatory networks controlling growth and developmental processes such as seedling photomorphogenesis, photoperiodic regulation of flowering, shade avoidance and responses to biotic and abiotic stresses [8–13].
Plant BBX proteins are characterized by the presence in the N-terminus of one single 40-residue B-box domain or two arranged in tandem. B-box domains are classified into two types, known as B-box1 (B1) and B-box2 (B2) depending on their consensus sequence and the distance between the zinc-binding residues [7]. Some members of the BBX family have an additional domain, termed CCT (CONSTANS, CO-like, TIMING OF CAB1: TOC1), composed of 42–43 amino acids located in the C-terminus [7, 8]. While the B-box domain is involved in mediating transcriptional regulation and protein-protein interaction, the CCT domain plays important functions in transcriptional regulation and nuclear protein transport [8]. In addition to these domains, some BBX proteins contain a valine-proline (VP) motif of six amino acids preceding the CCT domain. The BBX family consists of 32 members in Arabidopsis, 30 in rice and 29 in tomato which are classified into five groups based on the presence and number of B-box and CCT domains [7, 13, 14]. In Arabidopsis, groups I and II display two B-box domains and one CCT domain, group III includes one of each domain, group IV possesses two B-box domains and group V only one B-box domain [10].
Transcriptional analysis of several BBX genes in Arabidopsis revealed circadian-dependent regulation of expression [15]. Currently, the best characterized plant BBX protein is CONSTANS (CO, AtBBX1) in Arabidopsis. It belongs to group I functions as a transcriptional regulator of the expression of the Flowering Locus T (FT) gene encoding a “florigen” signal, FT, that triggers flower differentiation under long-day conditions (LD) [16–18]. Conversely, other proteins including AtBBX2 (CO-Like1, COL1) and AtBBX3 (COL2) also belonging to group I do not affect flowering time, but are associated with circadian regulation [19]. Other representatives of groups I and II like AtBBX4 (COL3) and AtBBX7 (COL9) are negative regulators of CO and FT gene expression, respectively [10, 20, 21]. The current knowledge regarding the function of group-III BBX proteins in Arabidopsis is very limited in plants. One representative, AtBBX16 (COL7), acts as an enhancer of the shade avoidance response in conditions of low red light/far-red light ratio (R/FR) and promotes branching under high R/FR ratio [22]. Group-IV BBXs are mainly involved in regulation of photomorphogenesis. AtBBX20 (BZS1), AtBBX21 (STH2), and AtBBX22 (STH3) proteins specifically promote photomorphogenesis [23–27] whereas AtBBX18 (DBB1a), AtBB19 (DBB1b), AtBBX24 (STO), AtBBX25 (STH) suppress photomorphogenesis [15, 27–29]. These antagonist are associated with BBX abilities to interact with a main photomorphogenesis-promoting factor HY5 [30], with COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) and E3 ubiquitin ligase, a dark-dependent repressor of photomorphogenesis mediating degradation of BBX proteins and HY5 factor in the dark [31]. For instance, the interaction of AtBBX21 and AtBBX22 with HY5 enhances its transcriptional activity on target genes [23–25, 32]. In turn, direct interaction of AtBBX24 and AtBBX25 with HY5 suppress this activity due to the formation of inactive heterodimers [27]. Epistatic analyses on BBX proteins and COP1 have shown that BBX4, BBX20, BBX21 and BBX22 repress the COP1 function, whereas BBX24 and BBX25 trigger it [21, 23, 24, 26, 27, 29, 33]. In other respects, the interaction of BBX20, BBX24 and BBX25 proteins with COP1 addresses them to proteasomal degradation [21, 26], whereas BBX21 and BBX22 are recruited by COP1 into nuclear speckles [23, 24]. Note that some members of the group IV are involved in shade avoidance responses by mediating cell elongation [22, 33] but with opposite actions since, BBX19, BBX21 and BBX22 inhibit, and BBX18, BBX24, and BBX25 promote hypocotyl elongation under a low R/FR ratio [33, 34].
In the present study, we characterized the genes encoding the B-box proteins in potato with regard to their structural properties and expression patterns during the day/night phases, etiolation and de-etiolation. We identified 30 genes encoding B-box proteins in the potato genome (Solanum tuberosum, ssp. tuberosum, cv. Desiree), and classified them into the five structural groups based on the presence of the B-box and CCT domains. According to the time-of-day specificity of their expression during both light and dark phases, we distinguished five distinct types. Further revealed that the expression of eight StBBX genes is controlled by the circadian clock and that the transcripts abundance of eleven StBBX genes undergoes marked changes during etiolation and de-etiolation conditions.
Material and methods
Plant material, growth conditions
Solanum tuberosum L., cv. Desiree plants were propagated in vitro on solid MS medium at 20/15°C (day/night) under a 150 μmol photon m−2 s−1 PFD and a 14-h photoperiod, and then planted in soil and grown in a controlled environment chamber under a 250 μmol photon m−2 s−1 PFD and 14 h light (8:00–22:00) and 10 h dark conditions at 20/15°C, respectively.
For circadian experiments, one set of 2-week-old chamber-grown plants was transferred to continuous light for 2 days under a PFD of 250 μmol photon m−2 s−1, while a second set of plants was left in the chamber under the same PFD and 14-h photoperiod. Material was collected every 6 hours for 2 days.
For etiolation/de-etiolation experiments, one set of 2-week-old chamber-grown plants was transferred to darkness for 4 days, and then retransferred to 14-h light/10-h dark under a PFD of 250 μmol photon m−2 s−1. A second set of plants was left in the chamber under the same PFD and a 14-h photoperiod (8:00–22:00). Material from both sets of plants was collected at the following time points: 6:00, 7:55, 8:15, 9:00, 14:00, 20:00 and 14:00 on the following day. Harvested plant material was frozen in liquid nitrogen and stored at -80°C for RNA isolation. Two independent experiments and two independent plant samples for each experiment were used.
Gene family member identification
The 32 AtBBX protein sequences were used for initial identification of putative potato StBBXs. Preliminary results were found in the Ensembl Plants (http://plants.ensembl.org/) database. For confirmation, we used the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) database and the PSI-BLAST search tool with algorithm parameters: 500 maximum target sequences by p-value 0.05. To eliminate the probability of omission of any BBX protein in the potato genome, the conserved B-box domain sequence in identified StBBX proteins served as queries to search the database with PSI-BLAST. SMART (http://smart.embl-heidelberg.de/) and Pfam (http://pfam.sanger.ac.uk/), (E-value < 0.05) databases were used to confirm the presence of the B-box and CCT signature, respectively. Full-length cDNA, gene orientation, chromosomal position and protein characteristics were obtained from the Ensembl Plants database. In order to find out additional copies of genes located close to the tandemly arranged StBBXs, the coding sequence of identified genes was subjected to BLAST searches using potato pseudomolecule database v 4.04 (E-value < 0.05) [35].
Gene isolation and sequencing
To determine whether the StBBX genes identified by in silico analysis are functional, specific primers for each gene were designed within their coding sequence and the corresponding transcripts were cloned and sequenced (S1 Table). Total RNA was prepared from leaves of two-week-old Solanum tuberosum plants and the whole flowers of two-month-old plants grown under a 14-h photoperiod using TRIzol Reagent RT (MRC). Equal amounts of total RNA were treated with RNase-free DNase I (Invitrogen). First-strand cDNA was synthesized from 1 μg total RNA using oligo(dT)18 primer and 200 units of Maxima Reverse Transcriptase (Thermo Scientific) at 55°C for 30 min, according to the manufacturer's protocol. The resulting single-strand cDNA was amplified using specific primers for each BBX gene (S1 Table). The RT-PCR product was purified from 1% agarose gel using a Gel Extraction Kit (Qiagen) and cloned into the pGEM-T vector (Promega). Two microliters from each ligation were transformed into the XL1-Blue MRF’ E. coli strain according to the manufacturer’s instructions and selected on LB plates containing 50 μg/ml ampicillin, 2% X-Gal and 100 mM IPTG. Plasmid DNA were isolated from E. coli with a GeneJET Plasmid Miniprep Kit (Thermo Scientific) and sequenced. The sequencing results were aligned and confirmed with BLAST (blastx).
Sequence alignment, phylogenetic analysis and cis-element identification
The multiple sequence alignment of 30 StBBX proteins was built using MUSCLE in the MEGA 6.06 software with default parameters. The alignment was used as an input to construct the phylogenetic tree with 1000 bootstrap replicates. The alignment of conserved sequences of B-box 1, B-box 2 and CCT domains was generated using ClustalW in MEGA 6.06 and a phylogenetic tree for each domain was constructed using neighbor-joining algorithms with default parameters. The putative StBBX orthologs in tomato and Arabidopsis were identified using the Ensembl Plants (http://plants.ensembl.org/) database. The putative transcription factor binding sites of StBBX genes were identified in the genomic sequences within 1000 bp upstream from the start codon using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database.
Quantitative real-time PCR analysis
Total RNA was prepared from potato leaves using TRizol Reagent (Invitrogen) according to the manufacturer's protocol. Equal RNA amounts were treated with RNase-free DNaseI. The quantity and quality of RNA were analyzed in 1% agarose gel using an Experion automated electrophoresis station (Bio-Rad). RNA concentration was measured using a NanoDrop photometer (Thermo Scientific). The first DNA strand was prepared using the Maxima Reverse Transcriptase (Thermo Scientific). Real-time PCR was performed using the CFX96 Touch™ Real-Time PCR detection system (Bio-Rad, Hercules, USA) and the Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) for 30 BBX analyzed genes (S2 Table). Technical duplicates were performed using independent cDNA synthesis reactions. Each assay using gene-specific primers amplified a single product of the correct size with high PCR efficiency (90–110%). The analysis of fluorescence data was conducted using the CFX™ Software (Version 3.0, Bio-Rad, Hercules, USA). All qPCR analyses were normalized using the threshold cycle (C1) values corresponding to the reference genes (18S and EF-1-α) [36]. The normalized expression of target gene (ΔΔCq) was calculated as the relative quantity of the target gene normalized to the quantities of the two reference genes according to the manufacturer's software, where the denominator of the normalized expression equation is the geometric mean of the relative quantities of the two reference genes for a given sample. The values presented are the means of two technical replicates from two independent biological samples.
Results
Identification of the potato StBBX gene family
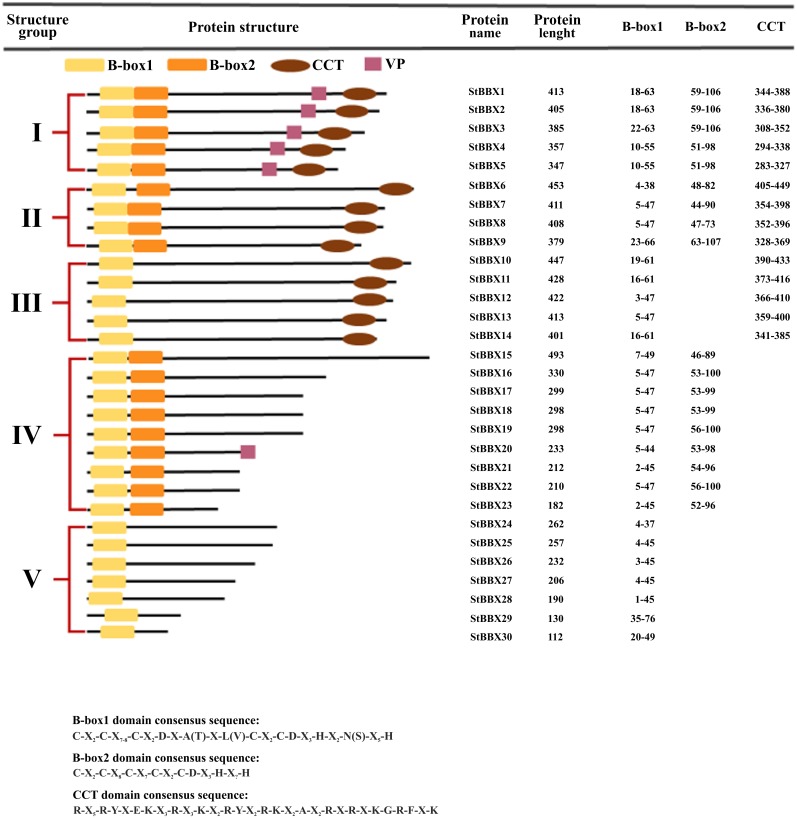
To identify B-box (BBX) genes in the potato genome, we searched ENSEMBL PLANTS and NCBI databases using the 32 Arabidopsis BBX sequences as queries. We identified 30 genes encoding BBX proteins with Gene IDs in ENSEMBL PLANTS, and among these only 28 had an accepted LOC number in the NCBI database. The presence of the B-box and CCT domains in identified potato sequences was confirmed using the SMART and Pfam databases, respectively. The consensus sequences of the B-box 1 and B-box 2 zinc finger domains are C-X2-C-X7-8-C-X2-D-X-A(T)-X-L(V)-C-X2-C-D-X3-H-X2-N(S)-X5-H and C-X2-C-X8-C-X7-C-X2-C-D-X3-H-X?-H, respectively. The consensus sequence of the CCT domain is R-X5-R-Y-X-E-K-X3-R-X3-K-X2-R-Y-X2-R-K-X2-A-X2-R-X-R-X-K-G-R-F-X-K (Fig 1). The identified genes were designated StBBX, followed by the Arabic numbers 1–30. The encoded StBBX proteins were classified into five structural groups based on the length, presence and number of characteristic B-box domains and the presence of the CCT domain and VP motif (Fig 1). Group I consists of five representatives, from StBBX1 to StBBX5, containing one double B-box domain, one CCT domain and the VP motif. Group II includes four proteins, from StBBX6 to StBBX9, exhibiting two B-box domains and one CCT domain, whereas proteins belonging to group III, StBBX10-StBBX14, have one domain of each type. Group IV, which includes two B-box domains, is most often represented with nine proteins (StBBX15-StBBX23), while group V is composed of seven members (StBBX24-StBBX30) containing only one B-box domain (Fig 1). Among all the identified StBBX genes, only two StBBX1 (originally designated StCO, AM888389) and StBBX20 (initially termed SsBBX24, ABC25454) were previously characterized in potato by Gonzalez-Schain et al. [37] and Kiełbowicz-Matuk et al. [11], respectively.
Fig 1. Structural classification of the StBBX family proteins.
The name and the length of corresponding StBBX protein and the position of characteristic B-box 1, B-box 2, CCT domains is shown on the right. The location and order of B-box 1, B-box 2 and CCT domains and VP motif within each protein is presented on the diagram.
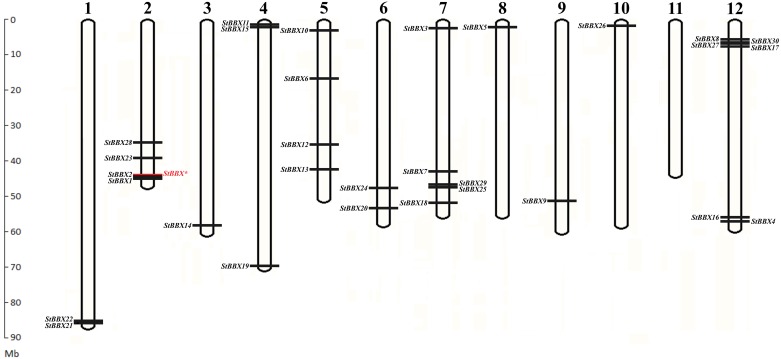
When searching through the ENSEMBL PLANTS database, we found that BBX genes are widely distributed throughout the potato genome, with the exception of chromosome 11 (Fig 2 and Table 1). Six of these are located on chromosome 12, five on chromosome 7, four on chromosomes 2 and 5, three on chromosome 4, two on chromosomes 1 and 6 and only one is located on chromosomes 3, 8, 9 and 10. In addition, six genes are located in the duplicated segmental regions of chromosomes 1 (StBBX21, StBBX22), 2 (StBBX1, StBBX2) and 12 (StBBX27, StBBX30). The detailed characteristics of StBBX genes concerning the gene ID, CDS, orientation, chromosomal position and protein products are shown in Fig 2 and Table 1.
Fig 2. Locations of 30 BBX genes on 12 potato chromosomes.
The scale on the left is in megabases. Chromosome numbers are indicated at the top of each bar. The gene names on the left and the right side of each chromosome correspond to the approximate locations of each BBX gene. An additional copy of StBBX* gene identified using potato pseudomolecule database v 4.04 was marked in red.
Table 1. The B-box gene family in Solanum tuberosum.
| Name | CDS length | Protein length | LOC (NCBI) | GENE ID | Gene position on chromosome | Gene orientation | Accession |
|---|---|---|---|---|---|---|---|
| StBBX1 | 1242 | 413 | CO CONSTANS | PGSC0003DMG402010056 | 2:45088023–45092647 | forward | KX576511 |
| StBBX2 | 1218 | 405 | LOC102598089 | PGSC0003DMG401010056 | 2:45098374–45101578 | forward | KX576512 |
| StBBX3 | 1158 | 385 | LOC102582832 | PGSC0003DMG400027475 | 7:2275710–2277328 | reverse | KX576513 |
| StBBX4 | 1074 | 357 | LOC102578495 | PGSC0003DMG400029365 | 12:58153725–58155118 | forward | KX576514 |
| StBBX5 | 1044 | 347 | LOC102585080 | PGSC0003DMG400026311 | 8:2052883–2054826 | reverse | KX576515 |
| StBBX6 | 1359 | 453 | LOC102603812 | PGSC0003DMG400025414 | 5:16834679–16838305 | reverse | KX576516 |
| StBBX7 | 1236 | 411 | LOC102580320 | PGSC0003DMG400017411 | 7:43163087–43172665 | forward | KX576517 |
| StBBX8 | 1227 | 408 | ND | PGSC0003DMG400028818 | 12:6077490–6085194 | forward | KX576518 |
| StBBX9 | 1140 | 379 | LOC102582339 | PGSC0003DMG400011378 | 9:52024565–52027713 | reverse | KX576519 |
| StBBX10 | 1344 | 447 | LOC102578395 | PGSC0003DMG400014566 | 5:2941260–2943570 | reverse | KX576520 |
| StBBX11 | 1287 | 428 | LOC102582864 | PGSC0003DMG400007749 | 4:1419075–1427624 | reverse | KX576521 |
| StBBX12 | 1269 | 422 | LOC102594696 | PGSC0003DMG400001263 | 5:42736399–42738599 | forward | KX576522 |
| StBBX13 | 1242 | 413 | LOC102603868 | PGSC0003DMG400005325 | 5:35571410–35577321 | forward | KX576523 |
| StBBX14 | 1206 | 401 | LOC102595945 | PGSC0003DMG400005633 | 3: 58929268–58930917 | reverse | KX576524 |
| StBBX15 | 1482 | 493 | LOC102587007 | PGSC0003DMG400005997 | 4:1835904–1840747 | forward | KX576525 |
| StBBX16 | 993 | 330 | LOC102590610 | PGSC0003DMG400029426 | 12:56979384–56982591 | forward | KX576526 |
| StBBX17 | 900 | 299 | LOC102593998 | PGSC0003DMG400019025 | 12:7848214–7852389 | reverse | KX576527 |
| StBBX18 | 897 | 298 | LOC102587821 | PGSC0003DMG400007061 | 7:52313046–52318239 | forward | KX576528 |
| StBBX19 | 897 | 298 | LOC102582815 | PGSC0003DMG400003711 | 4:70449684–70451592 | forward | KX576529 |
| StBBX20 | 702 | 233 | LOC102601030 | PGSC0003DMG400027017 | 6:53746105–53749420 | forward | KX576530 |
| StBBX21 | 639 | 212 | LOC102578169 | PGSC0003DMG400003109 | 1:86666771–86673688 | reverse | KX576531 |
| StBBX22 | 633 | 210 | LOC102593748 | PGSC0003DMG400030958 | 1:86524540–86526577 | forward | KX576532 |
| StBBX23 | 549 | 182 | LOC102606359 | PGSC0003DMG400003625 | 2:39712058–39715702 | forward | KX576533 |
| StBBX24 | 789 | 262 | LOC102580785 | PGSC0003DMG400026515 | 6:48085593–48086615 | reverse | KX576534 |
| StBBX25 | 774 | 257 | LOC102600093 | PGSC0003DMG400026181 | 7:47790598–47791812 | forward | KX576535 |
| StBBX26 | 699 | 232 | LOC102587352 | PGSC0003DMG400025024 | 10:1673524–1675193 | reverse | KX576536 |
| StBBX27 | 621 | 206 | LOC102585275 | PGSC0003DMG400013753 | 12:6868785–6869802 | forward | KX576537 |
| StBBX28 | 573 | 190 | LOC102603949 | PGSC0003DMG400022345 | 2:35131445–35132521 | reverse | KX576538 |
| StBBX29 | 393 | 130 | LOC102586344 | PGSC0003DMG400026169 | 7:47281684–47282147 | reverse | KX576539 |
| StBBX30 | 339 | 112 | ND | PGSC0003DMG400013178 | 12:6714370–6714828 | reverse | KX576540 |
To confirm the presence in S. tuberosum plants of transcripts corresponding to the 30 in-silico identified StBBX genes, we sought to obtain full-length cDNAs. To this purpose, we isolated total cellular RNA from leaves of two-week-old S. tuberosum plants grown under a 14-h photoperiod and collected at different time points in the light and dark phases. For cDNA analysis, the samples collected at different time points of the diurnal cycle were combined. RT-PCR analysis revealed the presence of products displaying lengths corresponding to those of genes identified by the in-silico approach. In the case of StBBX12, the presence of the highest amount of the corresponding transcript was detected in total cellular RNA isolated from the whole flowers of two-month-old S. tuberosum plants. The RT-PCR products were subcloned into the pGEM-T vector (Promega) and validated by sequencing. Sequencing data revealed that all the 30 isolated StBBX genes are identical with the genes annotated in the potato genome.
In order to show whether there are additional copies of BBX genes, we searched through the potato pseudomolecule database, and found one additional copy of the gene (with no annotation in the ENSEMBL PLANTS database) located proximally to the StBBX2 gene in the tandemly arranged StBBX1 and StBBX2 on potato chromosome 2 (Fig 2). This additional copy of the gene shares the highest identity with StBBX2 and StBBX1 (91.4% and 87.4% identity, respectively). Its putative coding region of 1234 nucleotides does not exhibit a full length of open reading frame, and encodes a truncated protein of 127 amino acid residues that is much shorter compared to the length of StBBX1 and StBBX2 which are comprised of 413 and 405 residues, respectively. The putative truncated protein comprises only the B1 and B2 domains flanked proximally by 20 amino acids and distally by 26 amino acids that allows it to be classified into structure group IV. Analysis of the expression of the putative BBX gene shows that the transcript corresponding to the fragment encoding the truncated protein was not detected in leaves of two-week-old Solanum tuberosum plants (data not shown). Taking into account the fact that this gene does not exhibit a full reading frame and its expression was not detected, it was not included into the list of the potato BBX genes (Table 1).
Phylogenetic analysis of the StBBX protein family
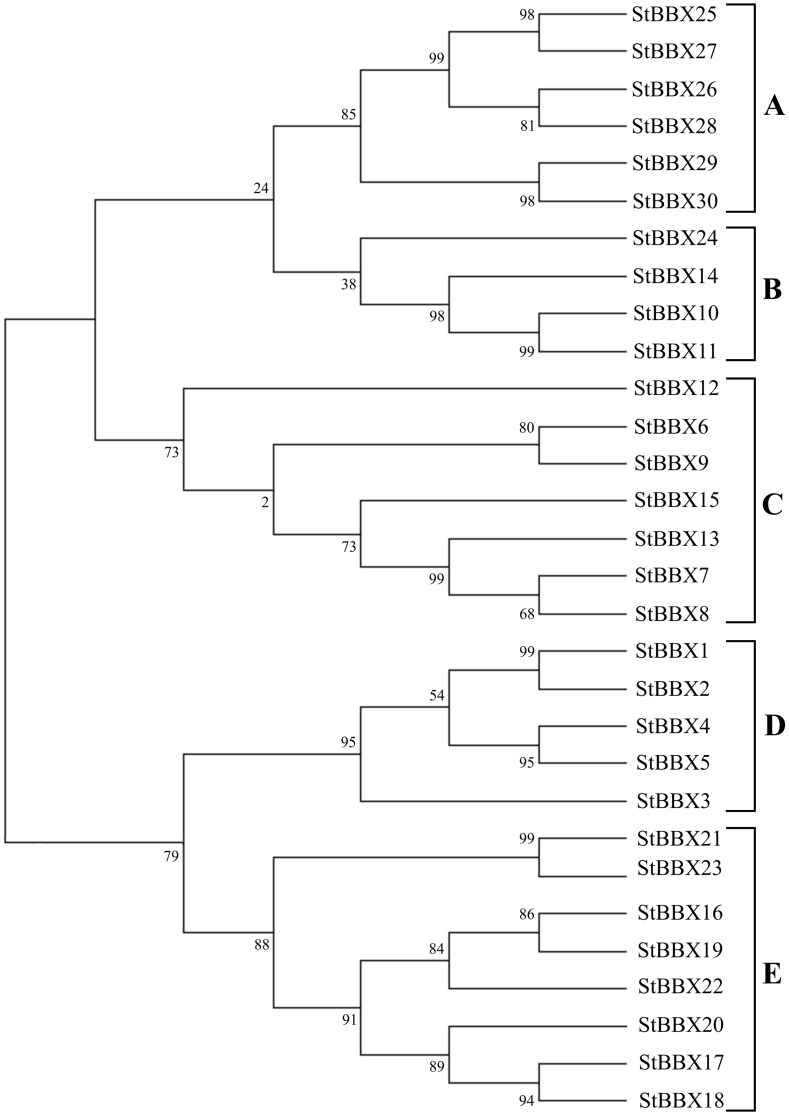
Based on the presence of the B-box and CCT domains and VP motif, the StBBX family proteins were divided into five groups (Fig 1), while the numbering of the proteins within a group was based on their length. To compare the structural classification with the phylogenetic relationships within the StBBX family, we performed a multiple full-length protein sequence alignment and constructed a neighbor-joining tree using MEGA 6.06 software. Based on the phylogenetic analysis, StBBX proteins were classified into five phylogenetic groups (A-E). The results revealed a considerable similarity between the groups defined from the structural features (I-V) and phylogenetic analysis (A-E), with the exception of phylogenetic groups B, C and E (Figs 1 and 3). According to the phylogenetic analysis, the StBBX24 protein classified into structure group V and is located in phylogenetic group B, while the StBBX12 and StBBX13 proteins from structure group III and the StBBX15 protein from structure group IV belong to phylogenetic group C (Fig 3). In addition, we performed other phylogenetic analyzes of the StBBX family based on the multiple alignments of B-box 1, B-box 2 and CCT domains. When aligning B-box 1 domains, we observed a similar phylogenetic relatedness among StBBX proteins compared to that revealed by alignment of full-length sequences (S1 Fig). However, in this classification, the StBBX24 protein localizes to group A (S1A Fig), and not to group B (Fig 3). Moreover, the divisions of the StBBX sequences with regard to the B-box 2 and CCT domains (S1B and S1C Fig, respectively) modify the classification based on the phylogenetic analysis of full-length sequences.
Fig 3. Phylogenetic analysis of the potato B-box family.
Full-length proteins of the 30 B-box members were aligned using MUSCLE in MEGA 6.06 software with default parameters [49]. The achieved alignment was used as a input to construct the phylogenetic tree with 1000 bootstrap replicates.
To identify the ortholog of potato BBXs in the tomato and Arabidopsis genome, we performed a BLAST search of the StBBX protein sequences using the Ensembl Plants database and found that orthologs of most B-box proteins in potato were identified in tomato and Arabidopsis (S3 Table).
Expression of the StBBX genes in the diurnal cycle
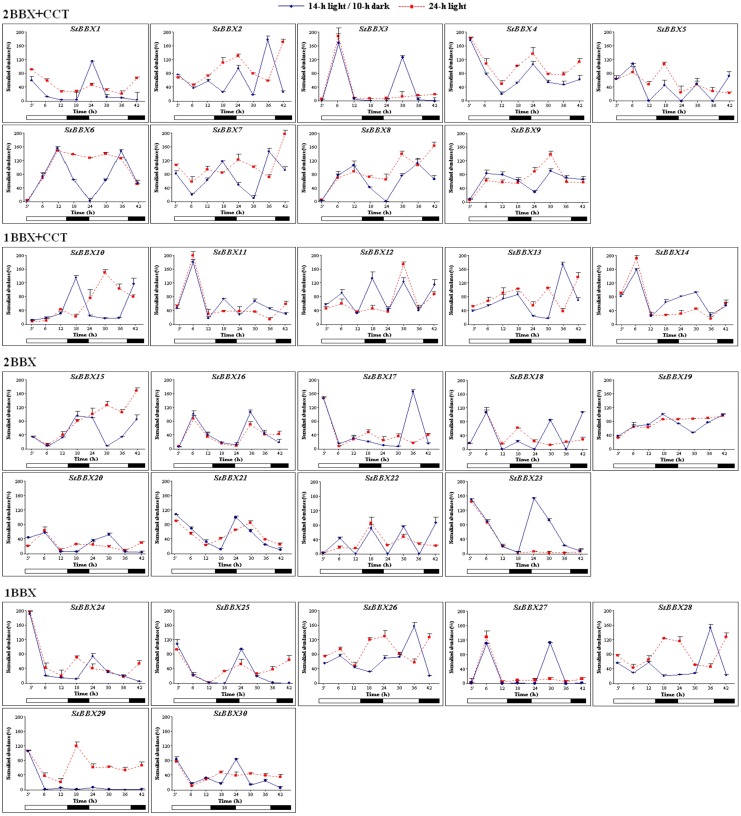
To determine which genes of the StBBX family undergo regulation as a function of the light-dark cycle, we first investigated the expression patterns of StBBX genes during the diurnal cycle (14h light/10h dark) for 42 h, at 6-hour intervals starting at the beginning of the light phase. As shown in Fig 4, most StBBX genes exhibited substantial variations in expression during the diurnal cycle. Interestingly, members belonging to each of the structural groups displayed different expression patterns in the light and dark phases. The diurnal changes in the transcript oscillation of StBBX genes between their maxima and minima were significant for most analyzed genes (Fig 4). We observed two main types of expression patterns, the first with maximal expression in the light phase and the second with maximal expression in the dark phase (Table 2). Furthermore, based on the timing of transcript levels we distinguished five different expression groups within these two types. The first and the second groups are light-specific with maximal expression at 6 and 12 hour of the light phase, respectively, while the third and fourth groups are specific for darkness with maximal expression at 4 and 10 hours in the dark, respectively (Table 2). Finally, when compacting the day-time points at the light/dark phase boundary and analyzing the expression of all StBBX genes under the light/dark cycle (Fig 5), an additional expression group, with maximal expression at 1 hour of light, was distinguished (Table 2). This expression group includes StBBX4, StBBX21, StBBX23, StBBX24, StBBX25, StBBX29 and StBBX30 genes.
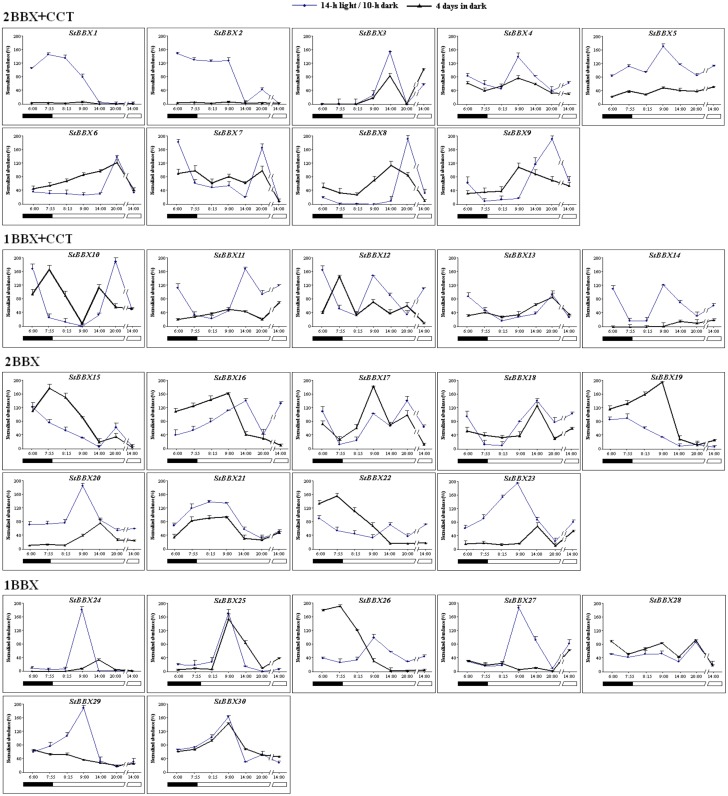
Fig 4. Diurnal and circadian regulation of StBBX family genes expression.
Analysis of StBBX transcript abundances in 2-week-old phytotron-grown S. tuberosum plants under a 14-h photoperiod (solid blue line) and continuous light (dashed red line) for 42 h during the subjective light and dark phases. Samples were collected at the indicated time points. qRT-PCR was performed as described in Material and Methods. To determine statistical significance between the maximum and minimum levels of the corresponding transcripts oscillation of the StBBX genes, we applied the Student's T-test.
Table 2. Classification of StBBX genes from Solanum tuberosum, cv. Desiree in terms of their expression in diurnal and circadian cycle and etiolation and de-etiolation conditions.
| Diurnal-regulated | Circadian-regulated | Etiolation specific | De-etiolation specific | ||||
|---|---|---|---|---|---|---|---|
| Light specific | Dark specific | ||||||
| max. expression at 1-h light | max. expression at 6-h light | max. expression at 12-h light | max. expression at 4-h dark | max. expression at 10-h dark | |||
|
StBBX4 StBBX21 StBBX23 StBBX24 StBBX25 StBBX29 StBBX30 |
StBBX3 StBBX9 StBBX14 StBBX16 StBBX18 StBBX20 StBBX26 StBBX27 |
StBBX6 StBBX8 StBBX13 StBBX17 StBBX28 |
StBBX10 StBBX15 StBBX19 |
StBBX1 StBBX7 |
StBBX1 StBBX4 StBBX9 StBBX14 StBBX16 StBBX20 StBBX21 StBBX25 |
StBBX16 StBBX22 StBBX26 |
StBBX6 StBBX8 StBBX9 StBBX15 StBBX17 StBBX19 StBBX20 StBBX23 |
Fig 5. Regulation of StBBX family genes expression during etiolation and de-etiolation.
Analysis of StBBX transcript abundances in 2-week-old phytotron-grown S. tuberosum plants under a 14-h photoperiod—control plants (solid blue line) and a continuous dark followed by a 14-h photoperiod (black bold line). Samples were collected at the indicated time points. qRT-PCR was performed as described in Material and Methods.
For the majority of StBBX genes, the changes in the expression profiles observed in the day/night cycle oscillate following a 24-h rhythm (Fig 4). However, large differences were observed between the genes with regard to the amplitude and distribution of maximal and minimal expression levels in the day/night cycle. Collectively, the oscillations were distributed throughout the 24-h cycle (Fig 4). For example, the transcript levels of StBBX3, StBBX16, StBBX18 and StBBX27 increased during the day and reached a maximum in the middle of the light phase, while the expression of others such as StBBX20, StBBX21, StBBX23, StBBX24, StBBX25 and StBBX30 increased during the night and reached the maximum at the beginning of the light phase (Fig 4). Otherwise, the expression of the StBBX7, StBBX10, StBBX15 and StBBX19 genes was induced in the middle of the light phase, then progressed throughout the subsequent hours, reaching a maximum in the night, and then decreased to the minimum in the first hours of the following light period (Fig 4). Taken together, these data indicate that the phase distribution of the transcript oscillation varied across the day and night; however, twenty StBBX genes displayed maximal levels of expression during the light phase.
Next, we investigated the expression of the StBBX genes in constant light for 42 hours. Upon continuous light, the expression profile of numerous StBBX genes was significantly modified compared to the 14/10-h day/night cycle in terms of the oscillation phase and amplitude. For instance, increased values of minima were noticed for StBBX2, StBBX5, StBBX6, StBBX9, StBBX10, StBBX28, StBBX29, and decreased values of the minimum for StBBX14 and StBBX25. An entirely disrupted rhythmicity of the diurnal expression patterns was observed for StBBX3, StBBX7, StBBX8, StBBX15, StBBX18, StBBX19, StBBX23, StBBX26, StBBX27, StBBX28, StBBX29 and an altered phase of cyclic expression by shortening or prolonging the total duration was noticed for StBBX10, StBBX13, StBBX21, StBBX24. The expression profiles of a few of the StBBX genes including StBBX1 and StBBX4 (structure group I), StBBX9 (structure group II), StBBX14 (structure group III), StBBX16, StBBX20, and StBBX21 (structure group IV), and StBBX25 (structure group V) were in synchrony under constant light with those observed during the standard day/night cycle (Fig 4), indicating that their diurnal expression is not directly regulated by light, but by the circadian clock (Table 2). Note that expression of StBBX3, StBBX23 and StBBX27 is totally abolished in the corresponding subsequent light phase upon continuous light (Fig 4), suggesting that diurnal oscillation of their expression requires a dark period.
Expression of the StBBX genes during etiolation and de-etiolation
The expression of StBBX genes was analyzed during etiolation and de-etiolation conditions in leaves of two-week-old S. tuberosum plants. As shown in Fig 5, the transcript levels of StBBX16, StBBX22 and StBBX26 were substantially higher at the end of the etiolation period compared to those in control plants, and this persisted at a similar level for approximately one hour in the subsequent light phase. Then, the transcript abundances dramatically decreased reaching the levels observed in control plants at the corresponding time-points in the light phase (Fig 5). These data showing increased transcript levels of StBBX16, StBBX22 and StBBX26 genes in continuous darkness suggest that these genes play physiological functions associated with etiolation processes. In contrast, the transcript abundance of StBBX6, StBBX8, StBBX9, StBBX15, StBBX17, StBBX19, StBBX20 and StBBX23 genes was very low by the end of the etiolation period, but dramatically increased in the subsequent hours of re-illumination (Fig 5), indicating that these genes could be involved in de-etiolation processes. Note that expression of the StBBX6 and StBBX8 genes in the control plants maintained under 14h light/10h dark conditions was induced after six hours in the light phase, much later compared to the timing of their expression in the control plants shown in Fig 4. Expression of the StBBX6 and StBBX8 genes might be very sensitive to any change occurring during the growth of the different sets of plants used for the subsequent experiments. Meanwhile, other StBBX genes exhibited widely disparate expression profiles during etiolation and de-etiolation compared to control conditions. Some genes, e.g. StBBX8, StBBX9, StBBX16 and StBBX19, displayed elevated transcript levels at the end of the fourth day of darkness, followed by a further increase within the following hours in the light, to reach levels similar to those observed in the control plants (Fig 5). Interestingly, the expression level of StBBX13 was low at the end of the etiolation period, but returned to the level observed in control plants with the onset of light; however, the transcript abundance of others, such as StBBX27 and StBBX29, did not increase in the subsequent light phase (Fig 5). In the case of StBBX3, StBBX21, StBBX25, StBBX28 and StBBX30 genes, there was no significant change in their expression profiles during the etiolation and de-etiolation processes (Fig 5).
Discussion
In the present study, we identified and characterized the potato StBBX gene family which is composed of 30 representatives with regard to structural properties and expression profiles under diurnal cycle (day/night phases) and during etiolation and de-etiolation conditions. Based on the presence and the characteristics of B-box and CCT domains and VP motif, StBBX genes were classified into five structural groups. On the other hand, based on the time-of-day specificity in expression patterns during light and dark phases, we distinguished several expression groups of StBBX genes, three specific for the light phase and two for the dark phase. The changes in the diurnal expression of several StBBX genes were shown to be under the control of the circadian clock. Moreover, we also demonstrated that the expression of numerous StBBX genes is differentially regulated during etiolation and de-etiolation conditions, indicating that they likely fulfill distinct functions in the diurnal regulation of developmental processes.
Previous phylogenetic analyzes of plant BBX proteins have been based on the genome sequences of A. thaliana and Oryza sativa, followed by comprehensive evolutionary analyzes of families from twelve other plant species by Crocco and Botto [8]. So far, 32, 30 and 29 BBX genes have been isolated and characterized in Arabidopsis, rice and tomato, respectively [7, 13, 14]. A uniform Arabidopsis BBX nomenclature was proposed by Khanna et al. [7]. As in Arabidopsis, rice and tomato, potato StBBX genes are also widely distributed throughout the chromosomes (Fig 2). The chromosomal localization of StBBX genes revealed that some genes were segmentally duplicated on chromosomes 1 (StBBX21, StBBX22), 2 (StBBX1, StBBX2) and 7 (StBBX27, StBBX30), while others were randomly distributed throughout the potato genome. Such a random distribution throughout the potato genome, including segmental duplication events, suggest that distinct mechanisms led to the diversification in the StBBX gene family, and that segmental duplication further contributed to an increase in the diversity of StBBX genes in potato. The lower number of BBX genes in potato genome compared to Arabidopsis may be due to the variable level of genome duplication in the two species. The structural classification of the StBBX proteins was largely consistent with the phylogenetic alignment of the full-length protein sequences, with the exception of four proteins: StBBX12 and StBBX13 belonging to structure group III were classified into phylogenetic group C, despite the lack of one B-box domain (B-box 2); while StBBX15 from structure group IV and StBBX24 from structure group V were classified into phylogenetic groups C and B, respectively, due to the lack of a CCT domain (Fig 3). The genes encoding these StBBX proteins probably lost the B-box 2 domain in evolutionary events, but retained the other common features of their structure group. To better understand the evolutionary origin of the B-box 1, B-box 2 and CCT domains in potato proteins, we performed alignments for each of the conserved domains followed by phylogenetic analyzes. Similar overall classification patterns were observed when B-box 1, B-box 2 and CCT domain sequences were used for the phylogenetic analysis; however, the classification of some StBBX proteins from structural groups II, III and IV has been changed. In addition, phylogenetic analysis of StBBX members enabled the identification of some orthologous genes in other species. For example, StBBX1 is an orthologous gene of Arabidopsis BBX1 (CO), while StBBX20 is an orthologous gene of Arabidopsis BBX24 (STO). Further, when searching through the potato pseudomolecule database, we identified an additional copy of the BBX gene on chromosome 2 located proximally to the StBBX2 gene of the tandemly arranged StBBX1 and StBBX2. This finding is in agreement with the data reported for the potato CONSTANS (CO) homologs [38]. These authors also identified three tandemly arranged homologs (StCOL1-StCOL3) on potato chromosome 2, and revealed that the StCOL3 transcript corresponding to the putative potato StBBX* gene (Fig 2) and to tomato SlCO2 has a deletion in the part encoding the CCT domain, and the transcript is rather undetectable [38].
With the exception of group V, the number of BBX members belonging to each structure group was different in potato and Arabidopsis (Fig 1 and data not shown). However, based on the presence of B-box1, B-box2 and CCT domains in Arabidopsis, rice and potato, we compared the number of proteins belonging to groups I-V and revealed variation between species in terms of the number of representatives in each group. Seven, nine and eleven proteins with two B-box and CCT domains were identified in rice, potato and Arabidopsis, respectively (Fig 1 and data not shown). The number of proteins with one B-box and CCT domain was 10 in rice, 5 in potato and 4 in Arabidopsis, while 10 proteins in rice, 9 in potato and 8 in Arabidopsis contained two B-box domains. The number of proteins with only one B-box domain was 3 in rice, 7 in potato and 7 in Arabidopsis [10, 13]. Based on these data, we suggest that the BBX genes from potato, Arabidopsis and rice have a common ancestor. However, differentiations occurred independently on the one hand following first the divergence of dicots and monocots, and on the other following that of Solanaceae and Brassicaceae.
As reported recently, numerous plant BBX proteins are implicated in multiple light-influenced processes, such as photomorphogenesis, flowering and shade avoidance [37, 39–43]. In Arabidopsis, the CO (AtBBX1) protein is involved in photoperiodic regulation of flowering under long-day (LD) conditions, but has no effect on flowering time under short-day (SD) [16, 18, 40]. Moreover, overexpression of another gene, COL5 (BBX6), causes early flowering in SD conditions in Arabidopsis [41], while overexpression of COL9 (BBX7) results in delayed flowering [20]. In rice, Heading date 1, the ortholog of Arabidopsis CO, shortens the time to heading [42], while OsCO3 is a floral repressor that regulates flowering under SD conditions [43]. In potato, StCO regulates photoperiodic tuberization in a graft-transmissible manner [37]. Chrysanthemum morifolium transgenic lines with suppressed expression of BBX24 flowers earlier than wild-type plants [44]. Given these observations, we determined the characteristics of the expression profiles of all the potato StBBX genes in terms of time-of-day and time-of-night specificity. Our results revealed a large diversity in expression patterns during the light and dark phases (Fig 4). The increased expression of numerous StBBX genes during the light phase suggests that they are regulated by components of light-signaling pathways, such as phytochromes and downstream regulators of photomorphogenesis, e.g. HY5, LAF1, FAR1, and PHY3 [45]. Other StBBX genes are specifically expressed during the dark phase indicating that they could be regulated by dark-specific factors such as Phytochrome-Interacting Factors (PIFs) that collaborate with phytochromes in the control of gene expression. On the other hand, some genes (StBBX2, StBBX11, StBBX13, StBBX14, StBBX26 and StBBX28) display changes in the amplitude of expression patterns during the diurnal cycle at the appropriate time points during 48-h, whereas the overall expression profile is unaltered. This amplitude variation of BBX gene expression suggests that post-transcriptional mechanisms contribute to the regulation of these genes. Our data suggest that BBX proteins participate in several developmental processes that are finely regulated as a function of the light and dark phases in the diurnal cycle. The fact that the light- and dark-specific expression groups are represented by members belonging to different structure groups indicates that all BBX types participate in these processes.
We also noticed that the diurnal expression of several StBBX genes is controlled by the circadian clock, indicating that these BBX proteins may function as components of circadian clock signaling (Fig 4). This finding is in agreement with the circadian regulation of other BBX genes reported for Arabidopsis BBX2, BBX3, BBX18, BBX19, BBX22, BBX24, and BBX25 [15, 19] and rice [13]. The light- and circadian-dependent expression of the StBBX genes is associated with the presence of several light-responsive and circadian-control regulatory elements in their promoter regions (S4 Table). The light-responsive elements may contribute to the regulation of StBBX gene expression in the light phase, while the B- and E-box variants of the G-box elements may contribute to the regulation of StBBX gene expression in the darkness. In turn, the presence of circadian regulatory elements in the promoters of some StBBX genes indicates that they can be controlled by the clock. Current understanding of the mechanisms controlling the expression of plant BBX genes is very limited. The best recognized mechanism has been described for the BBX1 (CO, CONSTANS) gene in Arabidopsis. CO functions as a transcriptional regulator of the expression of the Flowering Locus T (FT) encoding a florigen hormone FT that triggers flower differentiation under long-day conditions (LD) [17]. To induce the FT locus under specific day-length conditions, the timing of daily CO transcription and post-translational regulation of CO protein is precisely regulated [46]. As reported by Ito et al. [47], expression of the CO gene is activated by the FLOWERING BHLH (basic helix–loop–helix) proteins FBH1, FBH2, FBH3, but this is repressed in the morning by the CYCLING DOF FACTOR (CDF) proteins. It was recently revealed that the presence of the CIRC motif in the promoter region of circadian-regulated StBBX genes is necessary for the regulation of their expression, and that StZPR1 is a novel nuclear factor that binds the CIRC motif and regulates the circadian expression patterns of genes belonging to the B-box zinc finger family [48].
The light- and dark-specificity of the expression of some StBBX genes suggests that they are associated with those processes experienced during etiolation or de-etiolation. Indeed, etiolation-specific (StBBX16, StBBX22, StBBX26) and de-etiolation-specific (StBBX6, StBBX8, StBBX9, StBBX15, StBBX17, StBBX19, StBBX20 and StBBX23) genes have been distinguished (Fig 5 and Table 2). In Arabidopsis, the genes representing the group IV, such as AtBBX20, AtBBX21 and AtBBX22, have been shown to promote photomorphogenesis [9, 23–27], whereas AtBBX18, AtBBX19, AtBBX24, AtBBX25 and AtBBX32 suppress photomorphogenesis [27–29]. Our data complement those reported for Arabidopsis as they show that expression of the BBX genes classified to group II (StBBX6, StBBX8, StBBX9) is associated with de-etiolation in potato.
In conclusion, we have presented new data highlighting the fact that the expression of most StBBX genes is associated with the diurnal cycle; with two groups, this is specifically associated with the light or the dark phase. Furthermore, the expression of most StBBX genes oscillates over a period of 24-h, but the amplitude of oscillation and the phase distribution in respect of the light or dark phases are very specific to each gene. Expression of several StBBX genes is directly controlled by the circadian clock. Additionally, many StBBX genes are associated with etiolation and de-etiolation processes. These data clearly indicate that the StBBX genes are involved in diurnal regulatory networks of growth processes regulated by the light and dark, and the circadian clock.
Supporting information
The trees shown are based on the alignments of the protein sequences of the B-box 1 domain (A), B-box 2 domain (B) and CCT domain (C). The 30 B-box members were aligned using MUSCLE in MEGA 6.06 software with default parameters [49]. The achieved alignment was used as a input to construct the phylogenetic tree with 1000 bootstrap replicates.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors wish to thank Dr. Pascal Rey from CEA, DRF, BIAM, Laboratoire d’Ecophysiologie Moléculaire des Plantes, Saint-Paul-lez-Durance, France for critical reading of the manuscript. We thank Magdalena Biegańska from Institute of Plant Genetics PAS, Poznań, Poland for helpful assistance in preparing plant materials, and Agata Młodzińska from Genomic Laboratory, DNA Research Center, Poznań, Poland for help in bioinformatics analysis. We also thank Translmed (Cedar Hill, TX, USA), a proofreading and copyediting company, for help in copyediting and linguistic consultation regarding the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Science Centre under grant no. 2014/15/B/NZ9/04809. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McClung CR. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin Cell Dev Biol. 2013;24: 430–436. 10.1016/j.semcdb.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 2.Farré EM. The regulation of plant growth by the circadian clock. Plant Biol (Stuttg). 2012;14: 401–410. [DOI] [PubMed] [Google Scholar]
- 3.Kiełbowicz-Matuk A, Czarnecka J. Interplays of plant circadian clock and abiotic stress response network The Emerging Technologies and Management of Crop Stress Tolerance, Biological Techniques, ed. Ahmad P. and Rasool S. (Elsevier; ); 2014. pp. 487–506. [Google Scholar]
- 4.Gray WM. Hormonal regulation of plant growth and development. PLoS Biol. 2013;2: e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JH, Tsukaya H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot. 2015;66: 6093–6107. 10.1093/jxb/erv349 [DOI] [PubMed] [Google Scholar]
- 6.Bäurle I, Dean C. The timing of developmental transitions in plants. Cell 2006;125: 655–664. 10.1016/j.cell.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 7.Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T. The Arabidopsis B-box zinc finger family. Plant Cell. 2009;21: 3416–3420. 10.1105/tpc.109.069088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crocco CD, Botto JF. BBX proteins in green plants: insights into their evolution, structure feature and functional diversification. Gene. 2013;15: 44–52. [DOI] [PubMed] [Google Scholar]
- 9.Sarmiento F. The BBX subfamily IV: Additional cogs and sprockets to fine-tune light-dependent development. Plant Signal Behav. 2013;8: e23831 10.4161/psb.23831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19: 460–470. 10.1016/j.tplants.2014.01.010 [DOI] [PubMed] [Google Scholar]
- 11.Kiełbowicz-Matuk A, Rey P, Rorat T. Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum Double B-box gene. Ann Bot. 2014;113: 831–842. 10.1093/aob/mct303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson M, Staiger D. Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot. 2015;66: 719–730. 10.1093/jxb/eru441 [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Zhao X, Weng X, Wang L, Xie W. The rice B-Box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE. 2012;7: e48242 10.1371/journal.pone.0048242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu Z, Wang X, Li Y, Yu H, Li J, Lu Y, et al. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front Plant Sci. 2016;7: 1552 10.3389/fpls.2016.01552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, et al. The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2008;72: 1539–1549. 10.1271/bbb.80041 [DOI] [PubMed] [Google Scholar]
- 16.Robson F, Costa MW, Hepworth SR, Vizir I, Piñeiro M, Reeves PH, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28: 619–631. [DOI] [PubMed] [Google Scholar]
- 17.Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;87: 57–66. [DOI] [PubMed] [Google Scholar]
- 18.Valverde F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J Exp Bot. 2011;62: 2453–2463. 10.1093/jxb/erq449 [DOI] [PubMed] [Google Scholar]
- 19.Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001;26: 15–22. [DOI] [PubMed] [Google Scholar]
- 20.Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005;43: 758–768. 10.1111/j.1365-313X.2005.02491.x [DOI] [PubMed] [Google Scholar]
- 21.Datta S, Hettiarachchi GH, Deng XW, Holm M. Arabidopsis CONSTANS LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006;18: 70–84. 10.1105/tpc.105.038182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Zhang Z, Li H, Zhao X, Liu X, Ortiz M, et al. CONSTANS-LIKE 7 regulates branching and shade avoidance response in Arabidopsis. J Exp Bot. 2013;64: 1017–1024. 10.1093/jxb/ers376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-Box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007;19: 3242–3255. 10.1105/tpc.107.054791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, et al. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008;20: 2324–2338. 10.1105/tpc.108.061747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008;54: 205–219. 10.1111/j.1365-313X.2008.03401.x [DOI] [PubMed] [Google Scholar]
- 26.Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant. 2012;5: 591–600. 10.1093/mp/sss041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, et al. The Arabidopsis B-box protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell. 2013;25: 1243–1257. 10.1105/tpc.113.109751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holtan HE, Bandong S, Marion CM, Adam L, Tiwari S, Shen Y. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-Regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011;156: 2109–2123. 10.1104/pp.111.177139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M. Salt tolerance (STO), a stress-related protein, has a major role in light signaling. Plant J. 2007;51: 563–574. 10.1111/j.1365-313X.2007.03162.x [DOI] [PubMed] [Google Scholar]
- 30.Gangappa SN, Botto JF. The multifaceted roles of HY5 in plant growth and development. Mol Plant. 2016;9: 1353–1365. 10.1016/j.molp.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 31.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17: 584–593. 10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- 32.Chang CS, Maloof JN, Wu SH. COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 2011;156: 228–239. 10.1104/pp.111.175042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crocco CD, Holm M, Yanovsky MJ, Botto JF. AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 2010;64: 551–562. 10.1111/j.1365-313X.2010.04360.x [DOI] [PubMed] [Google Scholar]
- 34.Crocco CD, Holm M, Yanovsky MJ, Botto JF. Function of B-BOX under shade. Plant Signal Behav. 2011;6: 101–104. 10.4161/psb.6.1.14185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardigan AM, Crisovan E, Hamiltion JP, Kim J, Laimbeer P, Leisner CP, et al. Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell. 2016;28: 388–405. 10.1105/tpc.15.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005;56: 2907–2914. 10.1093/jxb/eri285 [DOI] [PubMed] [Google Scholar]
- 37.González-Schain ND, Díaz-Mendoza M, Zurczak M, Suárez-López P. Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 2012;70: 678–690. 10.1111/j.1365-313X.2012.04909.x [DOI] [PubMed] [Google Scholar]
- 38.Abelenda JA, Cruz-Oró E, Franco-Zorrilla JM, Prat S. Potato StCONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr Biol. 2016;26: 872–881. 10.1016/j.cub.2016.01.066 [DOI] [PubMed] [Google Scholar]
- 39.Yan H, Marquardt K, Indorf M, Jutt D, Kircher S, Neuhaus G, et al. Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 2011;156: 1772–1782. 10.1104/pp.111.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80: 847–857. [DOI] [PubMed] [Google Scholar]
- 41.Hassidim M, Harir Y, Yakir E, Kron I, Green RM. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta. 2009;230: 481–491. 10.1007/s00425-009-0958-7 [DOI] [PubMed] [Google Scholar]
- 42.Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SK, Yun CH, Lee JH, Jang YH, Park HY, Kim JK. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta. 2008;228: 355–365. 10.1007/s00425-008-0742-0 [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Ma C, Xu Y, Wei Q, Imtiaz M, Lan H, et al. A zinc finger protein regulates flowering time and abiotic stress tolerance in Chrysanthemum. Plant Cell 2014;26: 2038–2054. 10.1105/tpc.114.124867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeom M, Kim H, Lim J, Shin AY, Hong S, Kim JI, et al. How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis? Mol Plant 2014;7: 1701–1704. 10.1093/mp/ssu086 [DOI] [PubMed] [Google Scholar]
- 46.Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17: 75–86. 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 47.Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA. 2012;9: 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiełbowicz-Matuk A, Czarnecka J, Banachowicz E, Rey P, Rorat T. Solanum tuberosum ZPR1 encodes a light-regulated nuclear DNA-binding protein adjusting the circadian expression of StBBX24 to light cycle. Plant Cell Environ. 10.1111/pce.12875 [DOI] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The trees shown are based on the alignments of the protein sequences of the B-box 1 domain (A), B-box 2 domain (B) and CCT domain (C). The 30 B-box members were aligned using MUSCLE in MEGA 6.06 software with default parameters [49]. The achieved alignment was used as a input to construct the phylogenetic tree with 1000 bootstrap replicates.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.