Fig. 2.
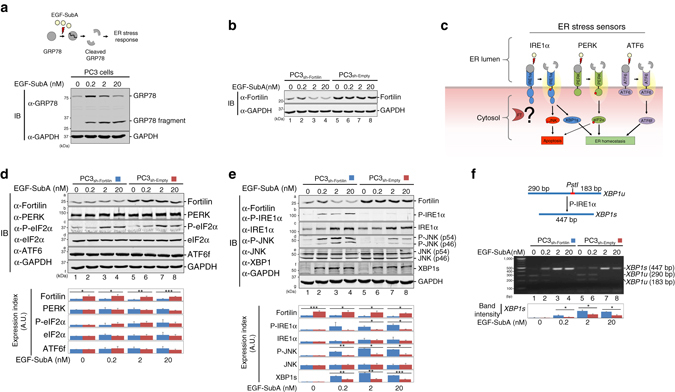
Fortilin selectively inhibits IRE1α signaling to protect against ER stress-induced apoptosis. a EGF-SubA dose-dependently cleaves GRP78 in PC3 cells. Wild-type PC3 cells were treated with the indicated concentrations of EGF-SubA, and their lysates were subjected to IB. b EGF-SubA treatment does not change the expression levels of fortilin in PC3 cells. PC3sh-Empty and PC3sh-Fortilin cells were treated with the indicated concentrations of EGF-SubA for 24 h and subjected to IB. c Three branches of ER stress signaling pathways. IRE1α, PERK, and ATF6 are the ER resident transmembrane proteins that signal ER stress when activated. IRE1α and PERK, but not ATF6, are known to induce apoptosis. d Fortilin does not regulate the PERK and ATF6 signaling pathways. PC3sh-Empty and PC3sh-Fortilin cells were treated with the indicated concentrations of EGF-SubA, and their lysates were subjected to quantitative IB for PERK, its downstream molecule eIF2α, and fragmented and activated ATF6 (ATF6f). Data were expressed as means ± s.d. (n = 3) and analyzed by two-tailed unpaired t-test. *P < 0.05; **P < 0.01; ***P < 0.005. e Fortilin inhibits the IRE1α ER stress signaling pathway. PC3sh-Empty and PC3sh-Fortilin cells were treated with the indicated concentrations of EGF-SubA and subjected to IB for IRE1α and its downstream molecules JNK and spliced XBP1 (XBP1s). P-IRE1α is the activated form of IRE1α, and P-JNK is the activated form of JNK. Data were expressed as means ± s.d. (n = 3, except for P-IRE1α and XBP1s where n = 2) and analyzed by two-tailed unpaired t-test. *P < 0.05; **P < 0.01; ***P < 0.005. f Fortilin inhibits the processing by IRE1α of XBP1 mRNA. PC3sh-Empty and PC3sh-Fortilin cells were treated with the indicated concentrations of EGF-SubA. The cDNA from total RNA from these cells was subjected to PCR and Pst1 restriction digestion (CTGCA|G) to semi-quantitatively evaluate the levels of XBP1s. Data were expressed as means ± s.d. (n = 2) and analyzed by two-tailed unpaired t-test. *P < 0.05. XBP1u unspliced XBP1
