Figure 5.
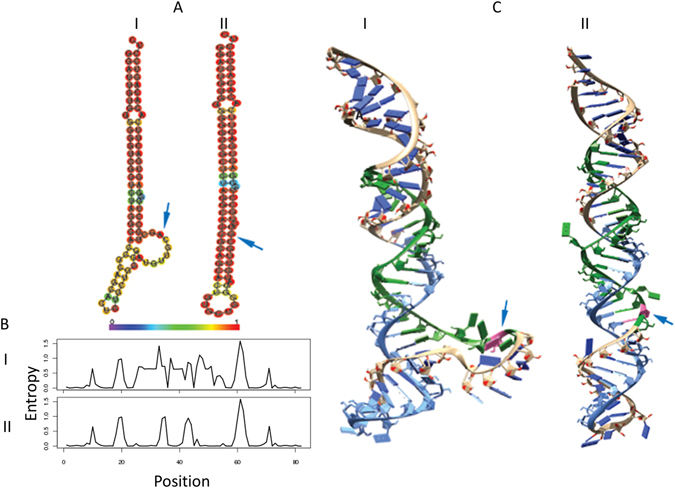
The two- and three-dimensional analysis for a representative pre-miRNA (mir-1301) unedited and edited sequence. (A) The minimum free energy secondary structures and its sequences, colored according to the base-pairing probability. The structure was predicted using RNAFold which uses a loop-based energy model and the dynamic programming algorithm. (B) The corresponding positional entropy for each position. (C) The lowest free energy three-dimensional structure was built using mcfold followed by mcsym where the RNAFold’s secondary structure information was given as structural constrain for unpaired nucleotides. The 5p arm is colored as blue while the 3p arm is colored green, with the edited nucleotide colored as pink. ‘I’ represent the unedited form while ‘II’ represents the edited form. The arrows mark the site of editing. The minimum free energy change at the two- and three dimensional levels for the unedited and edited pre-mirna-1301 is provided in Supplemental Table S5A.
