FIG 7.
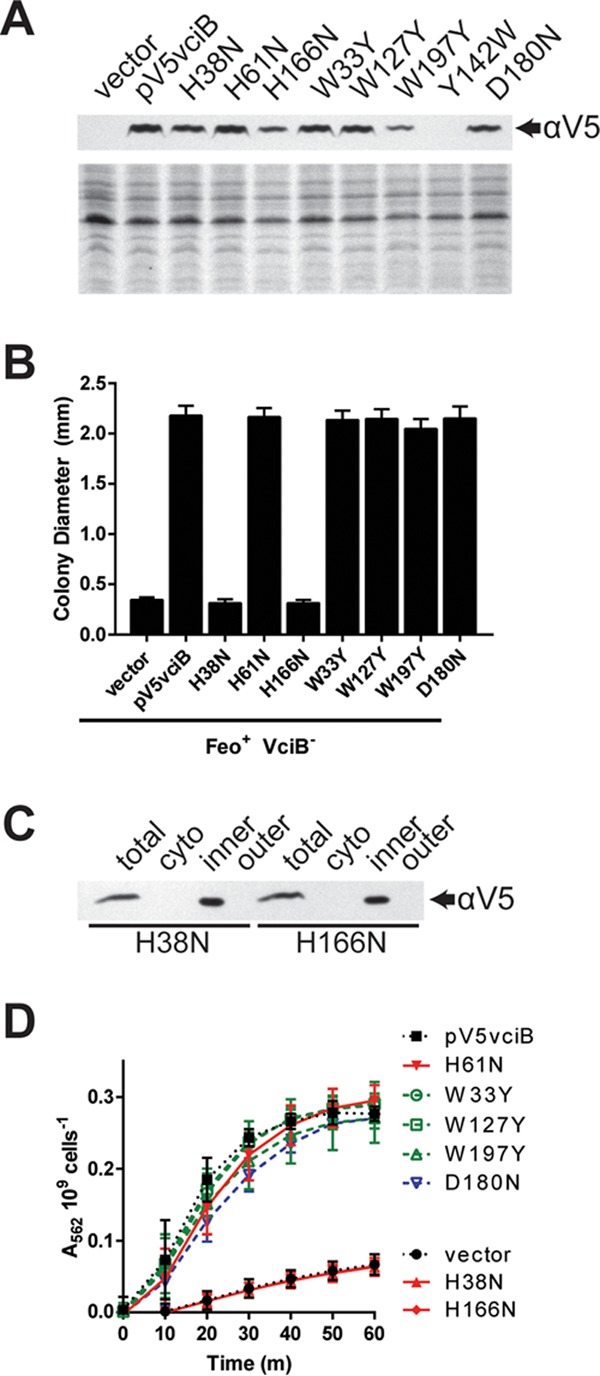
Identification of residues critical for VciB function. (A) (Top) Total cell lysates from EPV153 (Feo+ VciB−) bearing the indicated plasmids were probed for the V5 epitope. (Bottom) Coomassie blue-stained SDS-PAGE gel. (B) Point mutants generated in the V5-tagged VciB background were tested for the ability to complement the colony formation defect of the Feo+ VciB− strain (EPV153). Ten well-isolated colonies were measured using a reticle after 24 h of incubation at 37°C. The data are means and standard deviations and are representative of biological replicates. (C) Subcellular fractionation of cells with either V5-VciB H38N or V5-VciB H166N was used to determine localization. The total lysate (total), cytoplasmic (cyto), inner membrane (inner), and outer membrane (outer) fractions are marked. (D) Point mutants were tested for the capacity to reduce iron in the whole-cell iron reduction assay. H61N, W33Y, W127Y, W197Y, and D180N point mutants do not differ significantly from the wild-type pV5vciB sample; H38N and H166N mutants do not differ from the vector control. Significance was determined using a two-way ANOVA multiple-comparison test (Sidak).
