FIG 1.
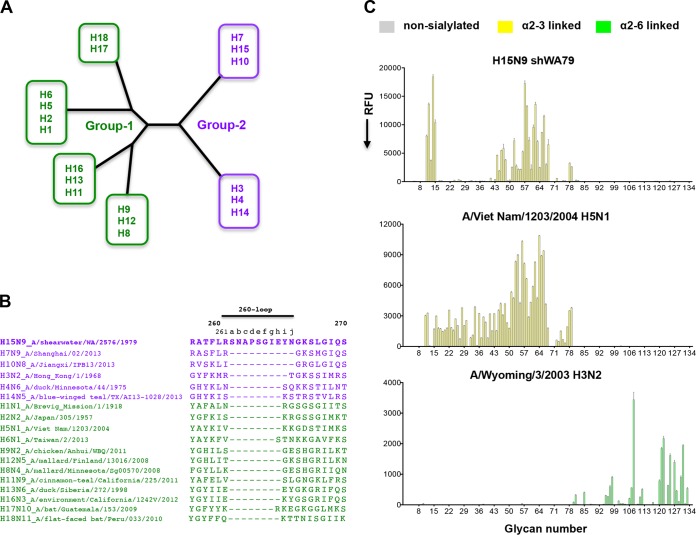
The amino acid sequence of the elongated 260-loop and the receptor binding specificity of the H15N9 shWA79 HA. (A) Schematic representation of the phylogenetic tree of influenza A virus HAs. The HAs are divided into group 1 and group 2, each of which can be further subdivided into subgroups. (B) Alignment of the HA 260-loop sequences of representative influenza viruses from all influenza A virus HA subtypes. The alignment indicates a marked elongation of the 260-loop of H15 HA compared to other HAs. The sequences of the group 1 and group 2 HAs are colored green and purple, respectively. (C) Glycan microarray analysis of the receptor binding specificity of recombinant H15N9 shWA79 HA indicates avian-type specificity and preferential binding of short, sulfated linear glycans (glycan numbers 11 to 15) and long, branched, O-linked and N-linked glycans. The A/Viet Nam/1203/2004 H5N1 and the A/Wyoming/3/2003 H3N2 HAs were used as controls for α2-3- and α2-6-linked sialoside binding, respectively. The mean signal (in relative fluorescence units [RFU]) and standard error were calculated from six independent replicates on the array. α2-3-linked sialosides are shown by yellow bars (glycans 11 to 79 on the x axis), and α2-6-linked sialosides are shown by green bars (glycans 80 to 135). Glycans 1 to 10 are nonsialylated control glycans and shown in gray. Glycans imprinted on the array are listed in Table S1 in the supplemental material.