Fig. 4.
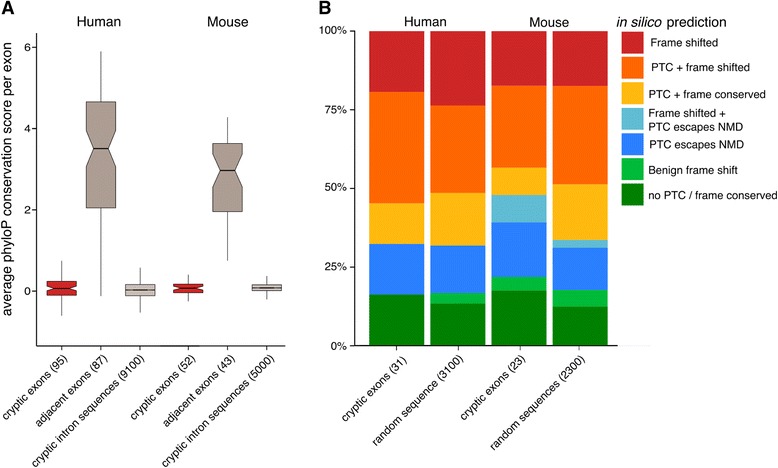
Conservation and premature termination codon analysis. a Per exon average PhyloP conservation scores in cryptic exons, adjacent exons within the same set of genes (when available) and randomly sampled sequences from the cryptic containing intron. Box plots show first quartile, median and third quartile with the notches representing the 95% confidence interval of the median. Whiskers represent the minimum and maximum values that fall within 1.5 times the interquartile range. b The functional impact of cryptic exon inclusion on the host transcripts in human and mouse. Colours indicate the category of prediction and box size indicates the proportion of the total group of exons in each category. Categories from top to bottom: Frame shifted (red); Frame shifted and at least one premature termination codon (PTC) included (orange); PTC included only (yellow); Frame shifted but the resulting PTC falls within 50bp of the exon-exon junction, creating a truncated protein of novel sequence (light blue); PTC falls within 50bp of the exon-exon junction, creating a truncated protein (dark blue); Frame shift without any PTCs to create a protein of novel sequence (light green); Cryptic exon inclusion does not frame shift nor introduces a PTC, behaving as a benign cassette exon (dark green). For each species there is a corresponding set of predictions calculated from random sequences with matching lengths
