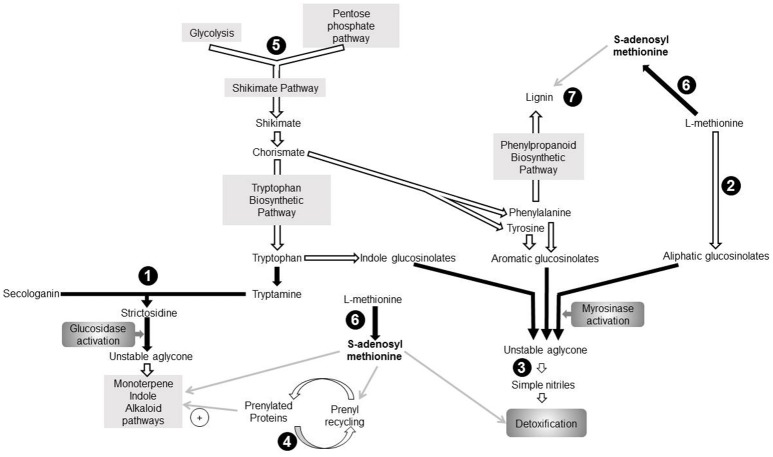
Figure 3.
Seven proteins identified in the aging leaf lipid droplet (LD) core proteome participate to secondary metabolism pathways. Enzymes identified in leaf LDs are represented in black circles. Pathways are represented by gray rectangles. Black arrows indicate direct reactions, white arrows represent indirect reactions, and gray arrows depict involvement of one component in a pathway. (1): 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAHP; AT1G22410), (2): Strictosidine synthase (AT1G74010; AT1G74020), (3): Farnesylcysteine lyase (AT5G63910), (4): Methionine adenosyltransferase3 (AT2G36880), (5): 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein (AT2G25450), (6): Nitrile specifier protein5 (AT5G48180), (7): Peroxidase superfamily protein (Peroxidase70; AT5G64110). DMAPP, Dimethylallyl diphosphate; IPP, Isopentenyl diphosphate; MEP, Methyl-D-erythritol 4-phosphate; MIA, Monoterpenoid indole alkaloid biosynthetic pathway; PP, Diphosphate.