Fig. 1.
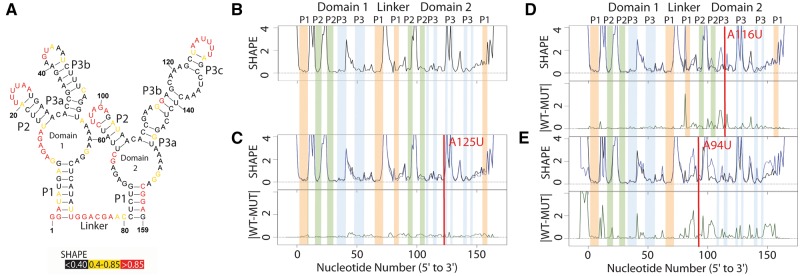
Structure change patterns in SHAPE trace data for the glycine riboswitch aptamers from Fusobacterium nucleatum. (A) Published WT structure for the apo glycine riboswitch aptamers consistent with the crystal structure and multiple independent structure probing experiments (Butler et al., 2011; Kladwang et al., 2011a,b,c). Red nucleotides indicate high SHAPE reactivity, yellow indicates mid-range reactivity, and black indicates low reactivity. (B) The individual WT trace is shown in black; the colored bars indicate the structural regions for each of the aptamers: P1 (orange), P2 (green) and P3 (blue). (C) The WT trace (black) is overlaid with the mutant SHAPE trace (dark blue), and the absolute difference between the WT and mutant traces is below (dark green). A red bar on the traces shows the mutation site. The A125U mutation is a mutation that leads to no appreciable differences in structure. 100% of experts that classified this mutant labeled it as a non-changer. (D) The A116U mutation leads to a local structure change, where the mutant trace reactivity increases at the mutation site disrupting the P3 region of domain 2. 66% of experts that classified the A116U mutant labeled it as a local changer. (E) The A94U mutation leads to a global structure change, where the mutant trace reactivity increases at both the mutation site and at nucleotides distant in sequence space disrupting both the P1 and P2 regions of domain 2. 66% of experts that classified the A94U mutant labeled it as a global changer (Color version of this figure is available at Bioinformatics online.)