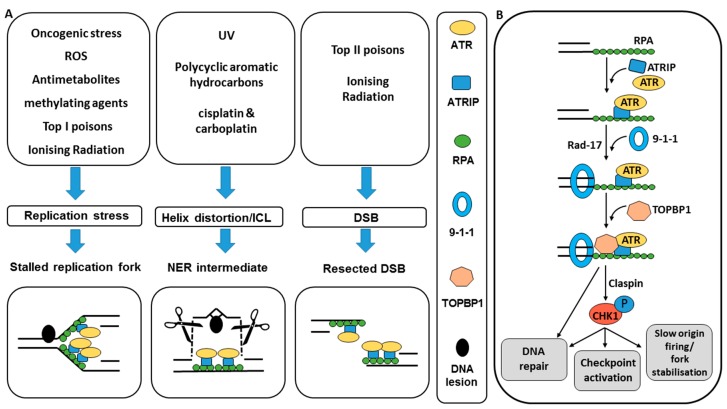
Figure 1.
Induction of ATR-CHK1 pathway activation and downstream signalling. (A) Anti-metabolites such as hydroxyurea (HU) and gemcitabine deplete the dNTP pool and cause stalled replication forks [13,14,15]. Topoisomerase 1 poisons and monofunctional DNA alkylating agents cause single strand lesions that can also stall/collapse replication forks. This increases replication stress and the availability of ss-DNA for ATR activation. Poly-cyclic aromatic hydrocarbon induced bulky DNA adducts and intra-strand cross-links resulting from agents such as UV and platinum based chemotherapy drugs are repaired by NER, leaving behind a short strand of ss-DNA [16,17]. The ssDNA is also present at the site of IR or topoisomerase II poison induced DSBs that have been resected by exo- or endo-nucleases [18]. In all cases the ssDNA is first coated by RPA. (B) RPA enables localisation of ATR to sites of DNA damage [10]. ATR recognition of the RPA-ssDNA complex is dependent on ATR-interacting protein (ATRIP). Localisation of the 9-1-1 complex via RPA interaction with RAD17 and subsequent recruitment of TOPBP1 and claspin leads to ATR activation and subsequent phosphorylation events leading to cell cycle arrest and DNA repair [9,15]. CHK1 acts as an intermediary in many of the DNA repair and DNA checkpoint reactions resulting from the activation of ATR and contributes to fork stabilisation and inhibiton of replication origin firing.