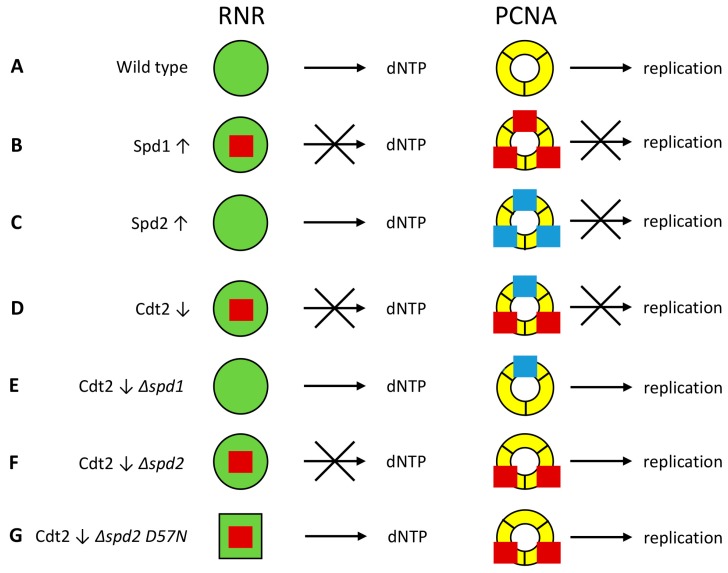
Figure 6.
Model for inhibition of DNA replication by Spd1 and Spd2. Spd1 (red squares) can inhibit ribonucleotide reductase (RNR, green circles), while both Spd1 and Spd2 (blue squares) can inhibit replication by binding to proliferating cell nuclear antigen (PCNA, yellow rings). S phase is inhibited if at least one of these two events occurs. (A) In wild type cells, both Spd1 and Spd2 are degraded, and hence dNTP production and elongation are not inhibited; (B) When Spd1 is overexpressed (↑), it inhibits both processes; (C) In cells overexpressing Spd2, only PCNA is inhibited; (D) In Cdt2 depleted cells (↓), Spd1 and Spd2 accumulate to a moderate level. Spd1 inhibits RNR, while both Spd1 and Spd2 are required to raise the concentration to a level where inhibition of PCNA can occur; (E) Deletion of spd1 relieves both types of repression; (F) When spd2 is deleted in Cdt2-depleted cells, repression of PCNA is lifted, but Spd1 still inhibits RNR; (G) The cdc22-D57N mutation changes the RNR configuration (green square) so that it can no longer be inhibited by dATP through negative feedback. Hence, sufficient amounts of dNTPs are formed even in the presence of Spd1 inhibition.