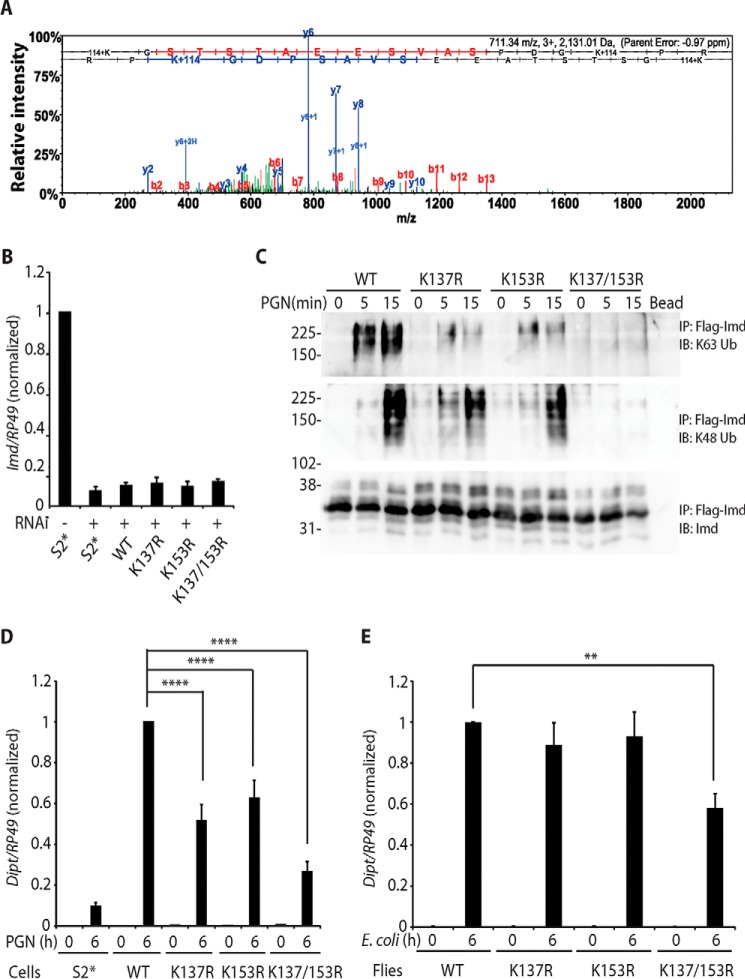
Figure 2.
Lys-137 and Lys-153 are the sites for Imd ubiquitin conjugation. A, Imd ubiquitination at Lys-137/Lys-153. Endogenous polyubiquitinated Imd was immunopurified and analyzed by tandem mass spectrometry. Ubiquitination sites were identified by their diglycine tags (K+114). A representative spectrum from two biologically independent mass spectrometric analyses is shown, with multiple diglycine-tagged peptides in each. B, Imd 3′-UTR targeting RNAi is ∼90% efficient. Parental S2* cell and stable cell line expressing Imd wild type (WT), K137R, K153R, or K137R/K153R were treated with dsRNA targeting Imd 3′-UTR. Endogenous Imd expression level was monitored by qRT-PCR with primers targeting Imd 5′-UTR. Data are shown as the mean ± S.D. from 3 independent experiments. C and D, inhibition of polyubiquitination and signaling by mutation of Lys-137 and/or Lys-153 in S2* cells. Stable cell lines expressing FLAG-Imd WT, K137R, K153R, or K137R/K153R were stimulated with PGN for indicated times. Endogenous Imd was knocked down by dsRNA targeting Imd 3′-UTR. Conjugation of FLAG-Imd was assayed by FLAG-immunoprecipitated (IP) and immunoblotting (IB) (C). The blot is representative of three independent experiments. Diptericin expression was monitored by qRT-PCR in D. Data are shown as the mean ± S.D. from three independent experiments. ****, p < 0.0001 (one-way ANOVA). E, inhibition of signaling by mutation of Lys-137 and Lys-153 in flies. An imd10191 (null) strain carrying transgenic WT, K137R, K153R, or K137R/K153R imd were stimulated with E. coli by septic infection at indicated. Diptericin expression was monitored by qRT-PCR. **, p < 0.01 (one-way ANOVA). Data are shown as the mean ± S.D. from three independent experiments.