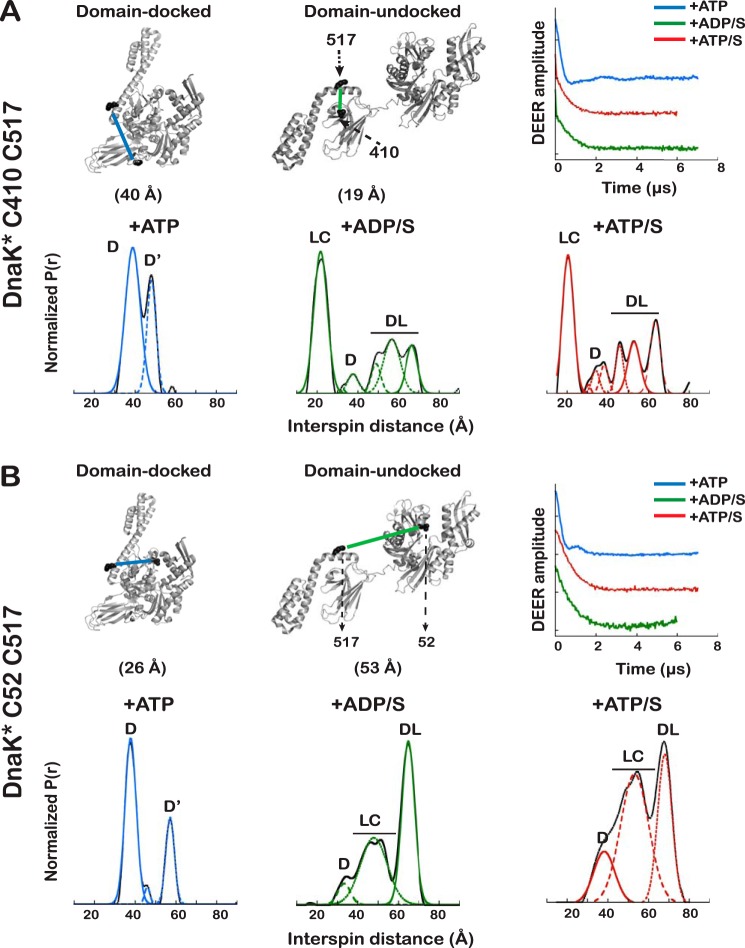
Figure 3.
DEER-derived distance distributions in DnaK* Cys mutants that report on dynamics of the α-helical lid. A and B, left, measured distance between spins in DnaK* Cys410-Cys517 and DnaK* Cys52-Cys517, respectively, based on Cβ–Cβ distance in PDB entries 2KHO and 4B9Q. The estimated distances do not consider spin-label conformers. Right, baseline-subtracted and normalized data showing time evolution of the interspin coupling. Bottom panels, interspin distance distributions for spin labels on DnaK* in the indicated ligand-bound states; solid lines, experimental data; dotted lines, fitted Gaussian components. Blue, ATP-bound; green, ADP/substrate-bound; red, ATP/substrate-bound. D, docked; U, undocked; DL, dynamic lid; LC, lid closed. Because of the higher uncertainty due to signal heterogeneity and dynamics, the distributions at mid-distances for the Cys52-Cys517 mutant were fit to only one Gaussian (three total components).