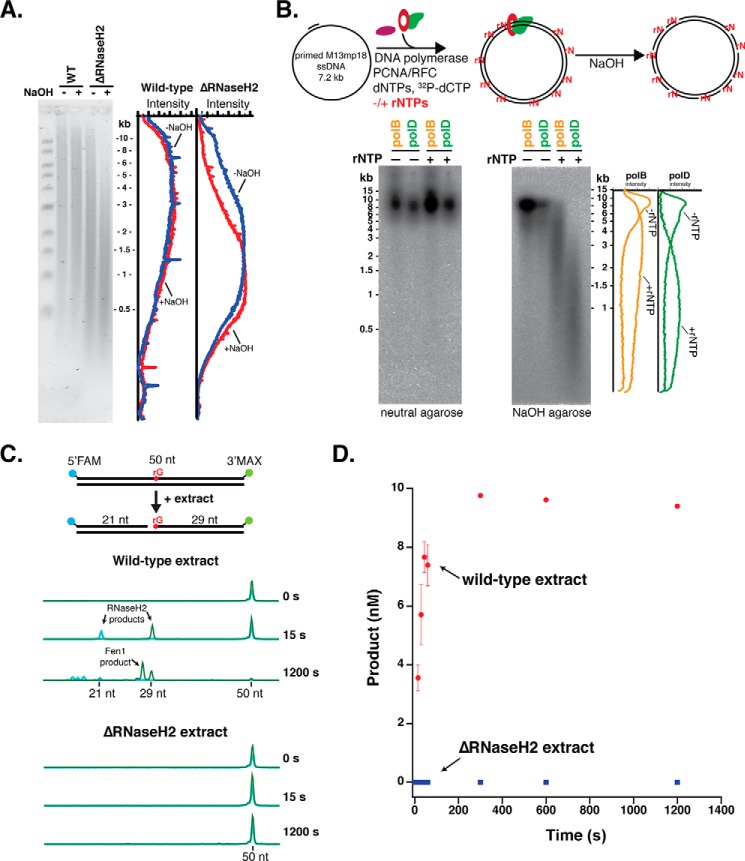
Figure 1.
Archaea require an RNaseH2-initiated RER pathway. A, genomic DNA from wild-type and ΔRNaseH2 T. kodakarensis (Tko) cells was treated with either 0.3 m NaCl or 0.3 m NaOH, separated on a 1% alkaline-agarose gel, and visualized using SYBR Gold staining. The fluorescence intensity distribution was quantified using ImageQuant software. B, primed M13mp18 ssDNA was fully extended by either Thermococcus sp. 9°N PolB or PolD with dNTPs or dNTPs/excess rNTPs (see “Experimental procedures”) with [32P]dCTP in place of dCTP for phosphorimaging. Full extension products were visualized by neutral agarose and rNMP incorporation was assessed by 0.3 m NaOH treatment and alkaline-agarose electrophoresis. C, RNaseH2 activity in Tko extracts was monitored by CE using a 50-bp dsDNA substrate with an embedded rGMP nucleotide, a 5′-FAM label, and a 3′-MAX label. Reactions were carried out over a time course from 15 s to 20 min at 60 °C. A subset of representative CE traces are shown indicating the formation of 21- and 29-nt RNaseH2 products. 3′-MAX-labeled Fen1 flap cleavage products (<29 nt) are also observed. D, the formation of the 29-nt MAX product was quantified over time for both wild-type (red) and ΔRNaseH2 (blue) extracts. Data are the average of three biological replicates and error bars indicate standard deviation.