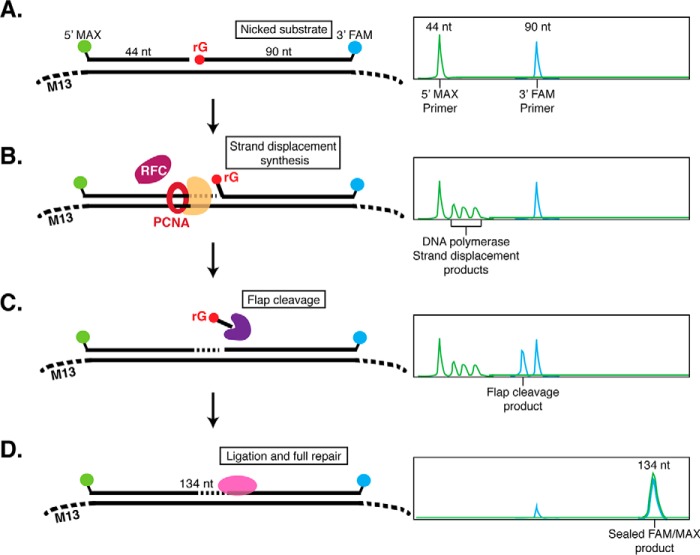
Figure 3.
Dual-label fluorescence assay to monitor post-RNaseH2 RER by capillary electrophoresis. A, the RER substrate was generated by annealing a 44-nt (5′-MAX labeled) oligonucleotide and a 90-nt (3′-FAM labeled with a 5′-phosphate-rGMP) oligonucleotide to ssM13 DNA. Annealed oligos form a DNA substrate containing a nick with 3′-OH and 5′-phosphate-rG termini to mimic DNA nicked by RNaseH2. On CE, the double-stranded DNA is denatured and ssDNA oligonucleotides can be visualized individually. On the right is a hypothetical CE trace representing the expected result with two individual MAX and FAM oligonucleotide peaks. B, when a strand-displacing DNA polymerase is added, 5′-MAX-labeled strand displacement products larger than 44-nt can be observed. C, when a flap endonuclease is added, 3′-FAM products smaller than 90-nt (predominantly by 1–2 nt) are observed. D, full sealing and repair by ligation results in a FAM- and MAX-labeled 134-nt product.