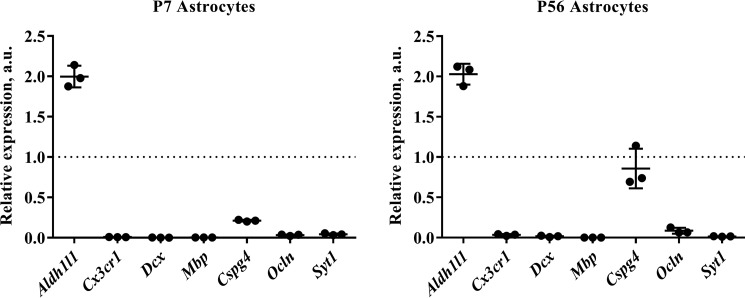
Figure 2.
Quantitative RT-PCR analysis of astrocytes isolated from young and adult mouse cortex. RNA was extracted from isolated astrocytes and reverse transcribed into cDNA. (As a positive control for relative quantification, RNA was also extracted from a single-cell suspension of whole cortex.) Quantitative PCR was then performed using a range of primers for cell type-specific markers, including Aldh1l1 (astrocytes), Cx3cr1 (microglia), doublecortin (Dcx) (neuronal precursors), myelin basic protein (Mbp) (oligodendrocytes), chondroitin sulfate proteoglycan (Cspg4) (oligodendrocyte precursors: NG2+ cells), occludin (Ocln) (endothelia), and synaptotagmin I (SytI) (neurons). Tata box-binding protein (Tbp) and cytochrome c1 (Cyc1) were used as housekeeping genes for normalization. (Full details of primer sequences used can be found in Table 1.) We routinely see a 2-fold enrichment of Aldh1l1 signal relative to control and a substantial decrease in contaminating cell types at both of the developmental ages we assayed, indicating that our astrocytes are of exceptional purity. The one exception was chondroitin sulfate proteoglycan (Cspg4), which was detectable at high levels in astrocytes isolated from P56 mice. Graphs show results from three independent cell isolation experiments (with a minimum of two technical replicates performed for each sample). qPCR runs on cells from different isolation experiments were repeated more than 3 times with the same results. The dotted line represents the level of mRNA in the positive control (whole-cortex cell suspension). a.u., arbitrary units. Graphs are plots of average ± S.D. (error bars).