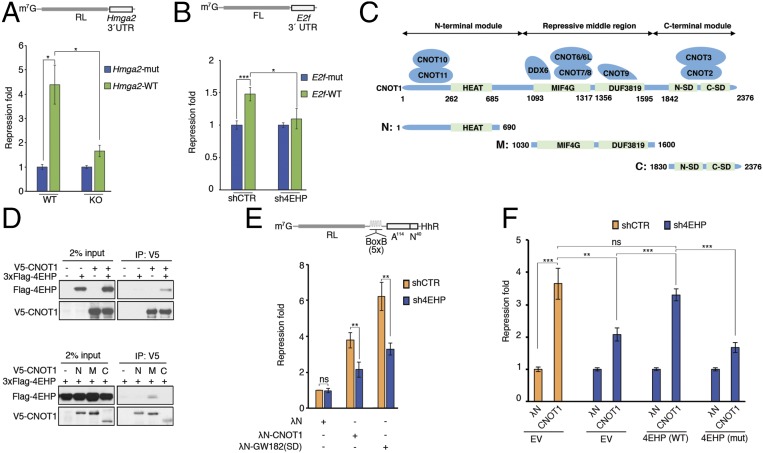
Fig. 2.
miRISC/CCR4–NOT-mediated translational silencing is impaired by 4EHP depletion. (A, Upper) Schematic representation of the RL-Hmga2 3′ UTR reporter. (A, Lower) WT and 4EHP-KO MEFs were cotransfected with RL-Hmga2 3′ UTR (WT) or mutant (Mut), along with Firefly luciferase (FL). Luciferase activity was measured 24 h after transfection. Renilla luciferase (RL) values were normalized against FL levels, and repression fold was calculated for the RL-Hmga2 3′ UTR (WT) relative to RL-Hmga2 3′ UTR (Mut) level for each population. The same data are shown as relative RL/FL levels in Fig. S2B. (B, Upper) Schematic representation of the FL-E2f 3′ UTR reporter. (B, Lower) shCTR and sh4EHP U251 cells were cotransfected with pGL3-FL-E2f 3′ UTR (WT) or a variant with mutations disrupting the two miR-17/20a–binding sites (Mut), along with RL. FL values were normalized against RL levels, and repression fold was calculated for the FL-E2f 3′ UTR (WT) relative to FL-E2f 3′ UTR (Mut) level for each population. (C) Diagram of full-length CNOT1 and the N-terminal, middle, and C-terminal fragments used in D. The binding partners in CNOT1 are also depicted and the main domains are shown in green. Amino acid positions at domain boundaries are indicated below the protein outlines. (D, Upper) Vectors expressing V5-CNOT1 and 3xFlag–4EHP (or control plasmid) were transfected into HEK293T cells. IP of V5-CNOT1 from RNase A-treated extracts was performed by using anti-V5 antibody. Purified proteins were analyzed by Western blot. (D, Lower) HEK293T cells were transfected with vectors expressing V5-CNOT1 fragments (as described in C): N, N-terminal region (amino acids 1–690); M, middle repressive module (amino acids 1,030–1,600); and C, C-terminal region (amino acids 1,830–2,376) and 3xFlag-4EHP. Extracts were subjected to anti-V5 IP, and the eluted fractions were analyzed by Western blot. (E) The 4EHP protein mediates translational repression by tethered CNOT1 and GW182(SD). (E, Upper) Schematic representation of the RL-5boxB-A114-N40-HhR reporter. Control HEK293T cells (shCTR) or cells depleted of 4EHP (sh4EHP) were cotransfected with vectors expressing either λN-CNOT1, λN-GW182(SD) or λN control, along with RL-5boxB-A114-N40-HhR or RL-A114-N40-HhR, and FL. RL luminescence was normalized against the FL level. (E, Lower) Fold repression of RL-5boxB-A114-N40-HhR relative to RL-A114-N40-HhR expression is shown. Repression of RL-5boxB-A114-N40-HhR by λN alone in shCTR cells was set as 1. (F) Rescue assay for λN-CNOT1–dependent silencing was performed, as described in E, in cells depleted of 4EHP. shCTR and sh4EHP HEK293T cells were transfected with the indicated plasmids in combination with constructs expressing shRNA-resistant versions of 3xFlag–4EHP (WT), 3xFlag–4EHPW124A cap-binding mutant (Mut), or empty vector (EV). The experiments illustrated in A, B, E, and F are represented as mean values (±SD) of three independent experiments. The P value was determined by two-tailed Student's t test. ns, nonsignificant. *P < 0.05; **P < 0.01; ***P < 0.001.