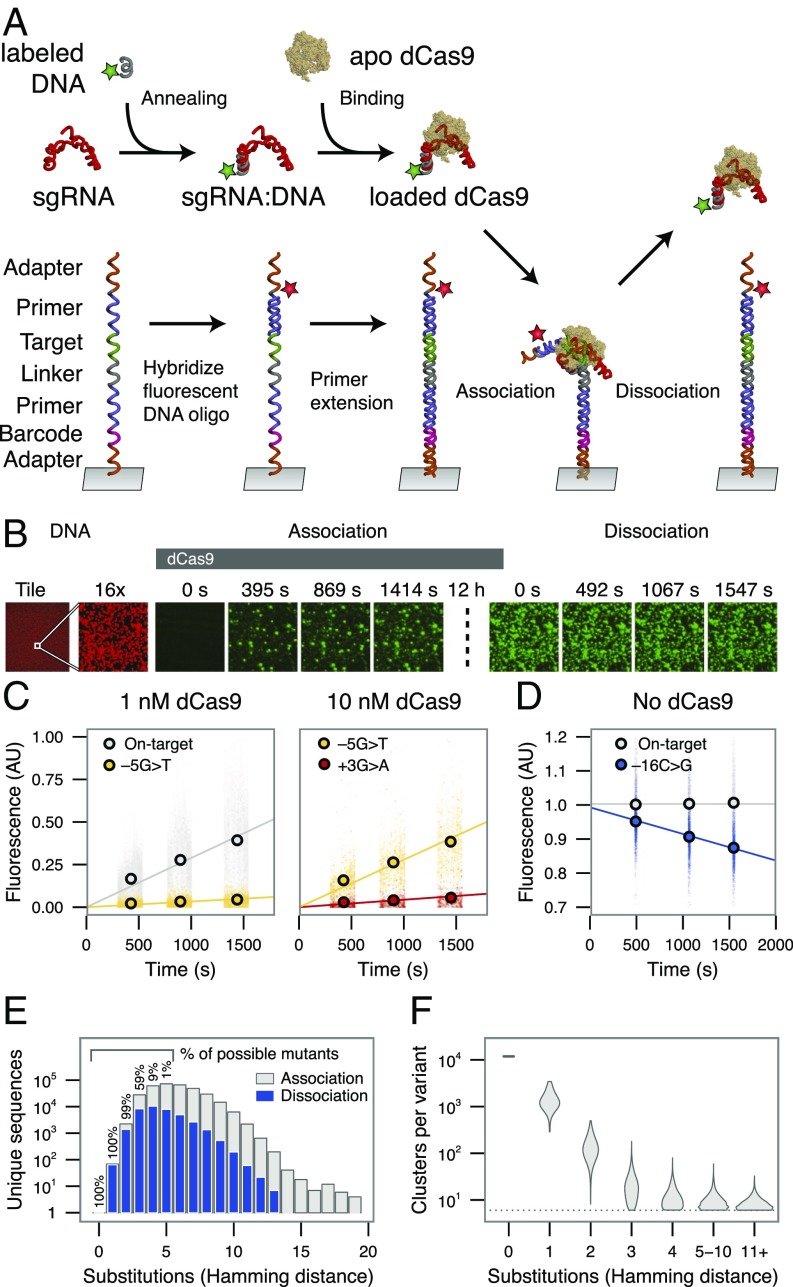
Fig. 1.
Quantifying dCas9 binding behavior on a massively parallel array. (A) Experimental procedure for high-throughput biochemical profiling. A fluorescent DNA oligo hybridized to the dCas9 sgRNA was loaded into the apo-dCas9. In parallel, an Illumina sequencing-compatible DNA construct was both labeled and made double-stranded by extending a second fluorescent oligo. dCas9 was flowed into the chamber, allowing association with double-stranded DNA. A dissociation experiment was then performed by quantifying the decrease in dCas9 signal upon dilution or chase. (B) Example images taken in two channels on the array, Alexa Fluor 647-labeled DNA (red) and Cy3-labeled dCas9 (green). A 12-h incubation, meant to saturate the clusters with dCas9, separates association from dissociation experiments (dotted line). For most clusters, signal accumulated in the on-rate experiment largely remains throughout the dissociation. (Magnification: right nine panels, 16×.) (C and D) Examples of (C) association and (D) dissociation lines fit to different targets. The +1 base refers to the first base of the PAM, −1 to the most PAM-proximal base, and −20 to the most PAM-distal base. (E) The total number (y axis) and percentage (in text) of possible targets profiled for each number of substitutions from the on-target site. Only a fraction of sequences with quantified on-rates are profiled for off-rates (blue) with high confidence. (F) Clusters per variant for targets with the given number of substitutions. AU, arbitrary units.