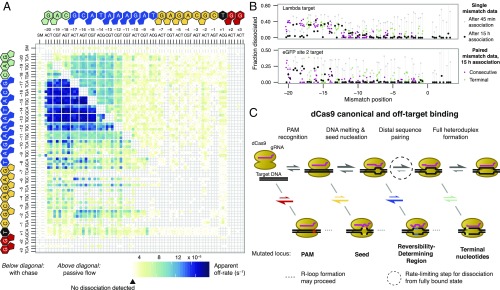
Fig. 3.
Variation in dCas9 dissociation rates suggests a model of Cas9 binding behavior. (A) dCas9 apparent off-rates for single and double mutants, as in Fig. 2B. Apparent off-rates are systematically higher in the presence of an unlabeled competitor dsDNA that prevents rebinding (below diagonal) than without (above diagonal). (B) Massively parallel filter-binding results for reversible binding at 10 nM dCas9. At shorter time scales (gray points), dissociation is most rapid for seed mutants for both targets. At longer time scales (all other points), dissociation is almost exclusively controlled by PAM-distal mismatches, either in isolation (black, positions –19 to –14) or when paired with a second mutation (green points). Consecutive mismatches in the seed show minimal dissociation after long association (pink points). (C) Model diagram for R-loop formation in different off-target contexts. Rates for protospacer mismatches are color-coded by off-target partition as in A. PAM and PAM-proximal mismatches affect early steps in the Cas9 target identification procedure, whereas distal mismatches influence later steps. Dissociation rates in A are likely products of the kinetics of unwinding of the R loop across the RDR. Dissociation rates associated with transient binding, as with most PAM mutants that fail to form R-loop structures, do not appreciably bind and thus are not captured in the dissociation experiment.