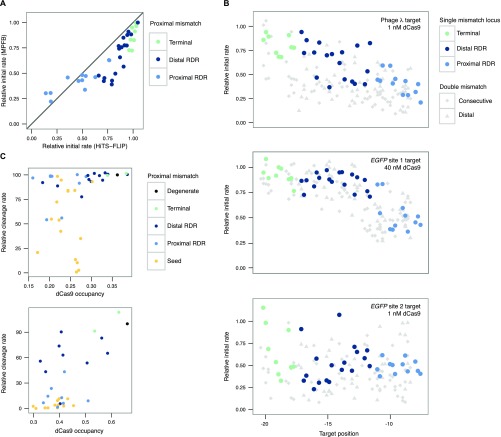
Fig. S9.
Insights from massively parallel filter binding data. (A) 1 nM data from both HiTS-FLIP and massively parallel filter binding experiments for the λ-target single mutants. The y = x line for perfect agreement is shown. Discordant points are enriched in regions with faster off-rates. (B) The relative initial rate, which we define as kobs times the fit max, plotted for each of the three target libraries at positions relevant for epistasis. Diamonds are double mismatches at consecutive target positions (e.g., a diamond at −18 represents mismatches at positions −17 and −18). Triangles are double mismatches where a proximal mismatch is paired with a distal mismatch at position −19 (e.g., a triangle at −15 represents mismatches at −15 and −19). The original lambda DNA target exhibits widespread negative epistasis. By comparison, the eGFP targets appear to exhibit both positive and negative epistasis. (C) dCas9 occupancy measured at single time points by massively parallel filter binding (10 nM dCas9) versus Cas9 cleavage measured in an eGFP fluorescence assay (25) for a mismatch-insensitive, slow-binding target (eGFP site 1, above), 15 h association, and for a seed mutant-sensitive, fast-binding target (eGFP site 2, below), 45 min association. Occupancy is correlated with reported rates, but seed mutants show lower cleavage rates than expected compared with RDR mutants.