Significance
The growing human population generates increasing demand for food and energy. Microalgae are a promising source of sustainable bioproducts whose production may not exacerbate worsening environmental problems. The green alga Chromochloris zofingiensis has potential as a biofuel feedstock and source of high-value nutraceutical molecules, including the carotenoid astaxanthin. We present a high-quality, chromosome-level assembly of the genome by using a hybrid sequencing approach with independent validation by optical mapping. Our analyses of the genome and transcriptome, in addition to experiments characterizing astaxanthin production, advance understanding of the green lineage and carotenoid production, and enhance prospects for improving commercial production of C. zofingiensis.
Keywords: Chlorophyceae, carotenoid biosynthesis, de novo genome, genome mapping, RNA-Seq
Abstract
Microalgae have potential to help meet energy and food demands without exacerbating environmental problems. There is interest in the unicellular green alga Chromochloris zofingiensis, because it produces lipids for biofuels and a highly valuable carotenoid nutraceutical, astaxanthin. To advance understanding of its biology and facilitate commercial development, we present a C. zofingiensis chromosome-level nuclear genome, organelle genomes, and transcriptome from diverse growth conditions. The assembly, derived from a combination of short- and long-read sequencing in conjunction with optical mapping, revealed a compact genome of ∼58 Mbp distributed over 19 chromosomes containing 15,274 predicted protein-coding genes. The genome has uniform gene density over chromosomes, low repetitive sequence content (∼6%), and a high fraction of protein-coding sequence (∼39%) with relatively long coding exons and few coding introns. Functional annotation of gene models identified orthologous families for the majority (∼73%) of genes. Synteny analysis uncovered localized but scrambled blocks of genes in putative orthologous relationships with other green algae. Two genes encoding beta-ketolase (BKT), the key enzyme synthesizing astaxanthin, were found in the genome, and both were up-regulated by high light. Isolation and molecular analysis of astaxanthin-deficient mutants showed that BKT1 is required for the production of astaxanthin. Moreover, the transcriptome under high light exposure revealed candidate genes that could be involved in critical yet missing steps of astaxanthin biosynthesis, including ABC transporters, cytochrome P450 enzymes, and an acyltransferase. The high-quality genome and transcriptome provide insight into the green algal lineage and carotenoid production.
The growing human population generates an increasing demand for food and energy, which intensifies environmental problems such as global climate change. Microalgae have the potential to become a major source of sustainable bioproducts, because they use solar energy, grow quickly, consume CO2, and can be cultivated on nonarable land (1, 2). However, there are presently considerable practical limitations in the production of biofuels from microalgae, resulting in low productivity and high costs. If microalgae can produce high-value bioproducts in addition to biofuel components, it could improve the economic viability of commercial algae production. The green microalga Chromochloris zofingiensis is a promising source of lipids for biofuel and the highly valuable ketocarotenoid astaxanthin, which has nutritive value because of its benefits in human health, making it a leading candidate for commercial scale-up (3–5). However, much remains unknown about the genome and regulation of metabolism in this alga.
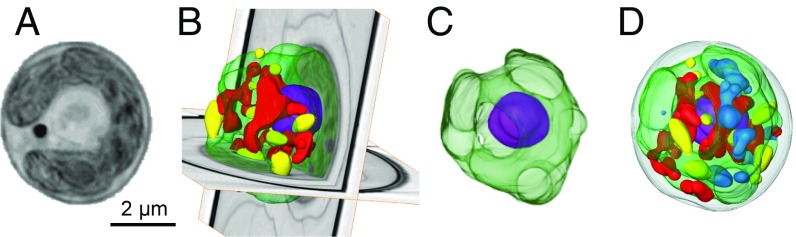
C. zofingiensis (division Chlorophyta, class Chlorophyceae, order Sphaeropleales) (6) is a simple ∼4-µm, unicellular, haploid, coccoid alga containing multiple mitochondria, which are visualized typically as a tubular network, and a single interconnected chloroplast that occupies ∼40% of the cell volume and contains starch granules (Fig. 1, Movies S1 and S2, and SI Appendix, SI Text). Most of the mitochondria are in close association with either the nucleus or the chloroplast. However, neither flagella (cilia) nor pyrenoids were visually observed. Because of the lack of obvious morphological characteristics, C. zofingiensis was originally described as a Chlorella species (6), at times transferred to the genera Muriella and Mychonastes, and finally placed using molecular sequencing into the genus Chromochloris (7). Similar to its close relative, the model alga Chlamydomonas reinhardtii, C. zofingiensis exhibits multiple fission with temporal separation between cell growth and cell division. C. zofingiensis primarily divides into two or four daughter cells (SI Appendix, Fig. S1 and Movies S3 and S4), but also can divide into 16 (SI Appendix, Fig. S1 and Movie S5), 32, or 64 cells (6). The regulation of cell division timing is unknown, but the daughter cells are the same size. Also like C. reinhardtii (8), the nucleus in C. zofingiensis divides before chloroplast division (SI Appendix, Fig. S1 and Movie S3). Intriguingly, C. zofingiensis has an extremely high photoprotective capacity compared with other algae and plants (9). Moreover, under specific conditions, C. zofingiensis can dramatically increase the production of lipids and secondary carotenoids (3–5, 10). This alga produces triacylglycerols (TAGs), the preferred lipid precursor for biofuel products and accumulates these lipids to some of the highest levels of 96 microalgae analyzed (3). Thus, C. zofingiensis is considered one of the most promising biofuel feedstocks for commercial production.
Fig. 1.
C. zofingiensis cell morphology. Cryo-soft X-ray tomography of a reconstructed cell with segmented nucleus (purple), chloroplast (green), mitochondria (red), lipids (yellow), and starch granules within the chloroplast (blue). (A) A representative orthoslice of the reconstructed cell. (See Movie S1 for transmission of single cell.) (B) Three-dimensional segmentation over two orthogonal orthoslices. (C) Segmented chloroplast and nucleus. (D) Fully segmented cell (see Movie S2 for rotating view).
Increased production of the highly valuable ketocarotenoid astaxanthin occurs in concert with accumulation of TAGs (4, 5). Astaxanthin has a broad range of commercial applications, including pharmaceuticals, nutraceuticals, cosmetics, food, and feed (11–13). Recent studies have highlighted the antioxidant and antiinflammatory benefits of astaxanthin for applications in human health including cancer, cardiovascular disease, neurodegenerative disease, inflammatory disease, diabetes, and obesity treatments (11, 12). Although astaxanthin can be produced synthetically, naturally produced astaxanthin is distinct in its esterification and stereochemistry (13–15). These differences result in natural astaxanthin having >20-fold stronger antioxidant activity than synthetic astaxanthin, and only natural astaxanthin has been approved for human consumption (14). Because C. zofingiensis is fast growing, can be cultured under many conditions (including with wastewater), and reaches high culture densities, C. zofingiensis has a higher potential to meet worldwide demand than other natural sources, such as the microalga Haematococcus pluvialis, yeast, transgenic plants, and crustaceans (13, 15–17). Thus, C. zofingiensis is a prime candidate to supply the world with natural astaxanthin and a source of renewable biofuel. However, improvements to maximize productivity and yield are needed, and key aspects of astaxanthin biosynthesis and regulation remain to be elucidated.
To better understand astaxanthin production and the biology of C. zofingiensis, we sequenced and assembled its nuclear, mitochondrial, and plastid genomes using a hybrid approach, constructed a transcriptome from 14 diverse conditions, examined transcriptomic changes through a shift from normal growth to that in high light, generated and analyzed astaxanthin-deficient mutants, and identified candidate genes involved in algal astaxanthin biosynthesis. The high-quality, chromosome-level genome assembly and accompanying transcriptome, combined with the capacity for genetic transformation (18), establish a molecular foundation to facilitate commercial development of C. zofingiensis and broaden understanding of the scope of metabolic and regulatory mechanisms found in the green lineage.
Results and Discussion
Whole-Genome Sequencing, Assembly, and Global Architecture.
For genome assembly of C. zofingiensis strain SAG 211–14, we used a hybrid approach blending short reads (Illumina), long reads (Pacific Biosciences of California), and whole-genome optical mapping (OpGen) (SI Appendix, SI Text and Datasets S1–S19, and refer to SI Appendix, Datasets Key). The combined power of these approaches yielded a high-quality haploid nuclear genome of C. zofingiensis of ∼58 Mbp distributed over 19 chromosomes (Fig. 2) in the tradition of model organism projects, as opposed to the fragmentary “gene-space” assemblies typical of modern projects using high-throughput methods and associated software. Approximately 99% of reads from the Illumina genomic libraries were accounted for, and nonplaceholder chromosomal sequence covers ∼94% of the optical map. Because no automated pipeline was found able to achieve the desired quality, methods are described in SI Appendix, SI Text.
Fig. 2.
C. zofingiensis nuclear genome. The assembled sequence of the 19 chromosomes of the nuclear genome is shown (top bar in each pair) with the matching restriction fragment length fingerprint from the optical map (bottom bar in each pair). Nominal plus strands run 5′ to 3′ left to right. Thin vertical divisions mark BamHI sites (in silico in top bars, optical consensus in bottom bars). Lines from restriction sites on one bar to another indicate a maximally scoring alignment computed with a dynamic programming algorithm similar to that used in OpGen’s MapSolver software. Black squares at chromosome edges indicate sequence assembly has reached telomere-associated repeats. Thick horizontal orange bars indicate explicit assembly gaps (runs of Ns). Thick horizontal yellow bars indicate additional known assembly issues as cataloged in Dataset S4. Light blue background shading shows where alignments are not one-to-one; shading is light green otherwise. Red dots mark possible (peri)centromeric loci. The end of chromosome 5 is discussed in SI Appendix, SI Text, and ∼24× copies of the rDNA unit likely predominate at the beginning of the large sequence gap at the end of chromosome 13. Unplaced contigs/scaffolds and 24 copies of the rDNA unit are shown near the bottom right.
We compared genome features of C. zofingiensis to four other green algae: C. reinhardtii, Coccomyxa subellipsoidea C-169, Chlorella sp. NC64A, and Monoraphidium neglectum (the closest relative with a sequenced genome), and the model plant Arabidopsis thaliana (SI Appendix, Table S1 and SI Text). Similar to most green algae, the genome of C. zofingiensis is about half the size of A. thaliana and C. reinhardtii; yet C. zofingiensis and all of the algae analyzed have more than double the number of chromosomes of A. thaliana. There is large variation in overall G+C content, with C. zofingiensis the most balanced (∼51% nuclear, 53% coding); although C. subellipsoidea C-169 is similar to C. zofingiensis, the other algal genomes have high G+C content and A. thaliana has low G+C content, and high G+C is associated with more fragmentary assemblies. Although Chlorella sp. NC64A has a large number of large regions with distinct G+C content, C. zofingiensis does not. Despite chromosome 5 appearing to end in a large (>1 Mbp) inverted repeat (discussed in SI Appendix, SI Text), the relative repetitive content of C. zofingiensis, like C. subellipsoidea C-169, appears to be low (∼6%). In contrast, M. neglectum is ∼50% higher and C. sp. NC64A ∼100% higher despite comparable genome sizes, and the large genomes of C. reinhardtii and A. thaliana are roughly double compared with the highest of the other four. After C. subellipsoidea C-169, C. zofingiensis contains the most repetitive fraction from novel repeats not known in Repbase Update (19), which presently focuses only on A. thaliana and C. reinhardtii. Gene density in C. zofingiensis is quite uniform over chromosomes, and there are no grand-scale gradients in genes or repeats such as found in, e.g., A. thaliana where each chromosome has several megabase pairs of pericentromeric heterochromatin (20), although some smaller scale repeat gradients are found near large assembly gaps and putative (peri)centromeres. RepeatMasker in conjunction with RepeatModeler and Repbase finds ∼5.0% of C. zofingiensis sequence consists of interspersed repeats [∼2.0% long interspersed nuclear elements (LINEs), ∼1.5% LTRs, ∼1.2% unclassified, and ∼0.4% DNA elements] with the remainder mostly simple repeats (∼1.0%) and with some satellites, low complexity sequence, and small RNA (total of ∼0.1%). The combination of smaller genome size, balanced G+C content, and low repetitive sequence fraction undoubtedly assisted assembly.
Complete (circular with no gaps or ambiguous nucleotides) mitochondrial and chloroplast genomes for C. zofingiensis strain UTEX 56 (formerly Bracteacoccus cinnabarinus) were already available as NCBI accession nos. KJ806268.1 (21) and KT199251.1 (22), respectively. We independently assembled equivalent complete genomes de novo for strain SAG 211–14 (SI Appendix, Table S1 and SI Text). The two strains were isolated from similar habitats in localities ∼300 km apart by different people in subsequent years. For the mitochondrial genome, the SAG and UTEX strains were resolved as 41,733 bp and 44,840 bp, respectively, with the same major protein-coding genes, tRNAs, and rRNAs in the same order (SI Appendix, SI Text) (21). However, a pairwise alignment exhibits only ∼66% nucleotide identity, with divergence concentrated intergenically and in rrnL4, where splicing differs. Restricted to coding sequence, nucleotide identity rises to ∼98%, and amino acid identity is ∼99% in translations under the National Center for Biotechnology Information (NCBI) Scenedesmus obliquus mitochondrial genetic code. For the chloroplast genome, the SAG and UTEX strains resolved as 181,058 bp and 188,935 bp, respectively, with a ∼6.7 kbp and ∼6.4 kbp, respectively, rRNA-related inverted repeat (SI Appendix, SI Text) (22). Neither the Illumina short reads nor Pacific Biosciences long reads were able to resolve the relative strand orientation of the two single copy regions for the SAG strain; a single contig was constructed with an arbitrary relative orientation that is opposite that given for the UTEX strain. Between the strains, all major protein-coding genes, tRNAs, and rRNAs are again the same in the same order. (In comparisons, the single copy regions were reoriented to agree.) Nucleotide identity is ∼83%, with divergence concentrated intergenically and with the largest single difference being a loss in the SAG strain of almost all of a ∼9.3-kbp UTEX region annotated as containing a ptz-like ORF. Coding sequence identity is ∼98%, and translation under the NCBI bacterial, archaeal, and plant plastid code gives ∼97% amino acid identity with lower identity in larger proteins (e.g., FtsH, RpoC2, and Ycf1). The low mitochondrial nucleotide identity was surprising given the presumed closeness of the strains.
The current C. zofingiensis assembly (“ChrZofV5”) successfully extended into telomere-associated repeats for 25 of 38 chromosome tips, and unplaced contigs appear to contain another 11 tips, leaving only two tips unaccounted. The canonical unit appears to be (CCCTAAA)n at 5′ ends [and the reverse complement, (TTTAGGG)n, at 3′ ends], similar to C. subellipsoidea C-169 and C. sp. NC64A and likely M. neglectum, although C. reinhardtii may prefer (CCCTAAAA)n. A comparison of counts of apparently telomere-associated reads vs. generic nuclear reads (and constraints imposed by the optical map) suggest an average of ∼3.5-kbp telomeric repeats per tip. Further, based on experience with particularly difficult sequences during assembly phases and analysis of the chromosomal distributions of specific dispersed and tandem repeat families, for most chromosomes exactly one region was identified as a putative (peri)centromeric locus. These loci are complex nested insertions of a ∼4.7-kbp circular consensus sequence that consists of a ∼4-kbp coding sequence of a type I/Copia LTR retrotransposon together with a ∼0.7-kbp spacer, and some 5S rDNA sequence (but apparently no large tandem arrays of a relatively short unit, such as in A. thaliana). The best NCBI BLASTX hits are to the filamentous green alga Klebsormidium flaccidum and the colonial green alga Volvox carteri. These regions are reminiscent of the Zepp clusters described in C. subellipsoidea C-169 (23), although the Zepp element is LINE-like and not of LTR type. Various analyses (including constraints imposed by the optical map) provided a rough estimate of only ∼25 kbp on average of (peri)centromere per chromosome in C. zofingiensis; perhaps construction of artificial chromosomes may be easier in C. zofingiensis than in some other organisms.
The canonical rDNA repeat unit of C. zofingiensis became apparent early in assembly because of its presence in relatively high copy number. It assembled as a 9,702-bp circular contig annotated by RNAmmer 1.2 as ∼6.6-kbp 28S followed by ∼1.1 kbp of spacer followed by ∼1.8-kbp 18S followed by ∼0.2 kbp of spacer. From the presence of homologous sequence on chromosome 13 leading into the large sequencing gap of that chromosome, the optical tandem repeat that begins that sequencing gap, and the presence of two BamHI sites in the consensus rDNA unit (creating alternating fragments of ∼6.0 kbp and ∼3.7 kbp that are consistent with the optical tandem repeat), it is estimated that ∼24× tandem copies of the rDNA unit predominate in the first ∼40% of the large sequence gap of chromosome 13. (Various analyses, e.g., SI Appendix, Table S1, assume 24 exact copies begin this gap.) The estimated number of copies is similar to M. neglectum, but drastically fewer than in the large genomes of A. thaliana and C. reinhardtii.
Genome Annotation and Transcriptomics.
To facilitate annotation, we generated a C. zofingiensis transcriptome by using RNA-Seq data collected from cells grown under 14 diverse conditions designed to capture a significant fraction of the cell’s transcriptional repertoire (SI Appendix, SI Text and Dataset S20). These conditions included treatments of different light intensities, nutrient limitations, and oxidative stress. Paired-end sequencing of transcriptome libraries was performed to facilitate determination of splice junctions, resolve close paralogous families, and de novo assembly (used as part of training the AUGUSTUS ab initio gene caller). To capture nonpolyadenylated transcripts, such as those from mitochondria and chloroplasts, libraries were prepared from total RNA depleted of rRNA.
RNA-Seq coverage, in conjunction with the de novo transcriptome assembly, was used to select a gene prediction method for producing gene models. Multiple pipelines, including Softberry’s Fgenesh, MAKER (24), and AUGUSTUS (25), were evaluated by using metrics such as RNA-Seq coverage capture and intron/exon boundary correlation with coverage. Of all evaluated pipelines, we selected AUGUSTUS as trained on the de novo transcriptome, which identified 15,274 nucleus-encoded protein-coding genes, of which 15,203 begin with a start codon (ATG) and end with a stop codon (TAA, TGA, TAG). The remaining 71 gene models are located on the unplaced contigs/scaffolds and likely extend beyond an edge in the assembly.
When the RNA-Seq libraries were aligned to the genome assembly, 95 ± 2% of reads aligned uniquely (mean ± SD, n = 10) and an additional 3 ± 1% aligned to multiple locations, suggesting that the genome assembly represents nearly all coding genes. Further, 55 ± 3% of RNA-Seq reads overlap by at least 80% with the coding portion of a gene model on the correct strand (mean ± SD, n = 10); only 1.3 ± 0.3% overlap with a gene model on the opposite strand. Current gene models do not include 5′ and 3′ UTRs; extending gene models 1 kbp upstream and downstream increases the percentage of reads aligning to the correct strand to 96 ± 2%.
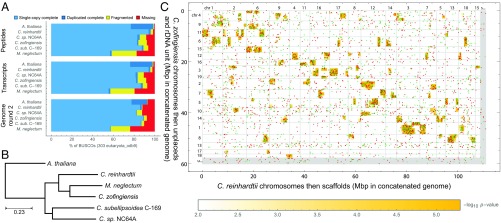
To further examine completeness, BUSCO (26) was run to identify C. zofingiensis orthologs for a set of 303 universal single-copy genes (USCOs) putatively universally found in eukaryotes as single copies (Fig. 3A). Given just the assembly, orthologs were identified by two-pass BUSCO for 92% of USCOs, with 97% of these declared complete. Given peptides, orthologs were identified for 92% with 91% complete. BUSCO analyses on the other organisms of SI Appendix, Table S1 suggest C. zofingiensis gene model quality is comparable to that of C. subellipsoidea C-169 and C. sp. NC64A, superior to M. neglectum, and inferior to extensively studied model organisms C. reinhardtii and A. thaliana. They also suggest the C. zofingiensis genome quality is higher than all of the other algae (including C. reinhardtii), with less fragmented and fewer missing orthologs.
Fig. 3.
Gene families. (A) BUSCO was run on the genomes (two-pass), splice variant-representative transcripts, and representative peptides of the six organisms of SI Appendix, Table S1 to locate copies of its set of 303 putative eukaryotic universal single-copy genes (USCOs). In each run, each USCO is classified as exactly one of single-copy complete, duplicated complete, fragmentary, or missing. Although the gene models of established model organism C. reinhardtii unsurprisingly fare better than the other algae, C. zofingiensis is comparable to the remaining algae, and its genome is the best of the algal genomes analyzed. A. thaliana’s excellent gene models show high duplication, suggesting BUSCO’s set of eukaryotic USCO’s are not all truly universally single copy. (B) Using a procedure based on reciprocal near-best global amino acid alignments, protein-coding gene families among the six organisms of SI Appendix, Table S1 were formed; a phylogram estimated from restriction to putative 1:1:1:1:1:1 orthologs is consistent with existing literature (27). (C) Scatterplots show scrambled syntenic blocks of conserved genes in the algal lineage (SI Appendix, Figs. S2–S10) (similar to fig. 2 in ref. 23). (Organism pairs involving the highly fragmented assembly of M. neglectum are omitted.) Each plot uses those gene families that, for the two organisms selected, have exactly one primary gene a in the first organism and exactly one primary gene b in the second organism. A dot with x, y coordinates at the midpoints of the span of the coding sequences for a, b is drawn in red if a and b are on the same nominal genomic strands and in green if they are on opposite strands; dots are plotted in a randomized order. Order of assembly sequences (but not nucleotides within sequences) is permuted on both axes so as to compact and emphasize statistically enriched regions (indicated by orange background shading); small inner numbers give relevant portions of the assembly’s sequence names for sequences at least 0.5 Mbp in length (e.g., “1” for “chr1”, i.e., “chromosome 1”). Rightward and downward are 5′ to 3′ on assembly plus strands and light gray lines mark assembly sequence boundaries; outer numbers give sequence scales (Mbp positions along concatenated genomes), which are equal horizontally and vertically. Small reference sequences (e.g., unplaced contigs labeled as “u…” and scaffolds labeled as “s…”) group at right and bottom sides (for which light gray dividing lines behind red and green orthologous gene dots may merge). Further details are given in SI Appendix, SI Text.
Before this work, NCBI contained 13 gene sequences marked as from C. zofingiensis. There was 99% or greater identity and 1% or fewer gaps as determined by BLAST alignment to our genome for 12 of the 13 (SI Appendix, Table S2). Only AY072815.1, heat shock protein 70, had limited identity, but this sequence was isolated from a different strain.
C. zofingiensis has the highest predicted coding sequence density (∼39%) of the SI Appendix, Table S1 organisms. The average length of its coding sequences (∼482 aa) is the longest apart from outlier C. reinhardtii, which helps bring C. reinhardtii to almost as high a fraction of coding sequence although its genome is much larger. However, the median length of C. zofingiensis coding sequences (∼347 aa) is more ordinary. The length of individual coding exons (whether by mean ∼291 bp or median ∼194 bp) of C. zofingiensis is the longest among the six, whereas the mean (∼5.0) and median (4) number of coding exons per gene is low, being more similar to M. neglectum and A. thaliana rather than the higher numbers seen in the other algae. The number of identified tRNA loci (75, and forming a complete set for the standard amino acids) is moderate like C. subellipsoidea C-169, rather than low as for C. sp. NC64A and M. neglectum, or high for the two large genomes of C. reinhardtii and A. thaliana.
To compare the C. zofingiensis proteome to others in the green lineage, we functionally annotated gene models by forming families of genes across the six organisms of SI Appendix, Table S1 using a method based on reciprocal near-best global amino acid alignments (SI Appendix, SI Text and Datasets S9–S18, and refer to SI Appendix, Datasets Key). This analysis generally permits one, many, or no genes per organism per family and separates genes into closer “primary” (putative orthologs) vs. further “additional” relationships (putative paralogs). The result contains 10,490 families involving more than one organism, of which 7,904 involve at most two genes per organism. There are some large families, with various histones constituting the largest. Approximately 73% of C. zofingiensis genes (and ≥∼60% of every genome) are placed in a family involving multiple organisms. All six genomes (including C. zofingiensis) show evidence of tandem duplication of genes. A phylogram (Fig. 3B) estimated from putative 1:1:1:1:1:1 orthologs placed C. zofingiensis closest to M. neglectum and then C. reinhardtii, forming a three-member clade that joins a two-member clade containing C. subellipsoidea C-169 and C. sp. NC64A, consistent with existing literature (27), and this whole-genome data analysis agrees with placing C. zofingiensis into genus Chromochloris (7).
Although we do not find large stretches of nucleotide synteny between C. zofingiensis and the other genomes, we do find among all members in the green algal lineage (except for M. neglectum, whose current assembly is too fragmented for such an analysis) highly significant genomically localized blocks of genes in putative orthologous relationships (Fig. 3C and SI Appendix, Figs. S2–S10), extending the result for C. subellipsoidea C-169 vs. C. sp. NC64A (fig. 2 in ref. 23). Whereas block boundaries are rather well defined, gene order and coding strands within blocks are generally completely scrambled. The topology of Fig. 3B can be reconstructed just from the pattern of syntenies. It is likely that the blocks represent random chromosomal rearrangements that accumulate over time and diverge after speciation.
To gain more insight into the metabolic function and cellular processes associated with specific proteins, we used in silico methods to predict subcellular localization of proteins encoded by the nuclear genome of C. zofingiensis. Using PredAlgo, an algal-specific subcellular localization prediction program trained on C. reinhardtii (28), we predicted nucleus-encoded proteins to distribute as ∼15% to the secretory system, ∼12% to the chloroplast, and ∼10% to mitochondria. The majority of proteins (∼63%) were predicted to be localized to other areas, which may be due to unidentified transit peptides, or the transit peptides of C. zofingiensis being significantly different from PredAlgo’s C. reinhardtii training set. Additionally, errors in gene models, especially in terminal regions, may result in inaccurate localization predictions. The predicted distribution is similar to what has been noted for C. reinhardtii (29).
Mitochondrial and chloroplast genes were highly expressed over a wide range of conditions (SI Appendix, Fig. S11). Despite the organellar genomes being significantly smaller and expressing many fewer genes, the transcripts expressed by the chloroplast and mitochondria represent a substantial portion of the total cellular mRNA. In an analysis of the transcriptomic data from 14 diverse growth conditions, 31 ± 9% and 7 ± 2% of total RNA-Seq reads uniquely mapped to the chloroplast and mitochondrion genomes, respectively (mean ± SD). Thus, RNA expression is dramatically higher for organellar genes: for the 73 protein-coding genes encoded in the chloroplast and 22 in the mitochondria, median transcript abundance across conditions was 686 and 419 FPKMs, respectively, in contrast to 5 FPKMs across all nuclear-encoded genes.
To identify genes that were more highly regulated under specific conditions, we compared expression of every gene over the 14 conditions and selected those with z-scores beyond ±2, plotting these as heatmaps (SI Appendix, Figs. S12–S21). The most prominent treatment to affect nuclear and plastid gene expression was oxidative stress by hydrogen peroxide (SI Appendix, Fig. S12), which significantly affected 3,934 genes. These genes were enriched for ABC-transporter domains (P = 1.0 × 10−6), suggesting that export of toxics and xenobiotics is a significant mechanism for handling environmental stress in C. zofingiensis. Similarly, singlet oxygen stress induced by the chemical Rose Bengal affected 1,477 genes (SI Appendix, Fig. S20) and heterotrophic growth on glucose identified 853 genes (SI Appendix, Fig. S14). Nutrient deprivation had similar effects on most genes and far fewer genes were identified by analyses; for example, only 21 genes were detected as highly enriched in the iron-deficient sample.
Cryptic Sex and Motility in C. zofingiensis.
Although C. zofingiensis has long been assumed to be asexual and nonmotile, we investigated the presence of putative cilia/flagella and meiosis genes in its genome via the computationally-identified gene families in conjunction with examination of associated gene expression across our conditions. The sequencing of the genome of C. sp. NC64A established a precedent for this type of analysis in green algae; similar to C. zofingiensis, neither sexual cycle nor flagella have been observed in C. sp. NC64A, yet its genome revealed meiosis-specific and primarily motile flagella genes, suggesting a cryptic sexual cycle (30). In the C. zofingiensis genome, we found putative orthologs for 73 of 78 genes (∼94%) in the CiliaCut (31), suggesting that there could be a previously unobserved motile life cycle stage with flagella in this organism. C. zofingiensis was missing only five genes: DLC4, FAP111, FBB5, IFT20, and Tctex1. (All gene symbols in this work are with implicit “[v5.2]” version suffixes.) In C. reinhardtii, the ift20 deletion mutant lacks flagella and is immotile (32), but perhaps C. zofingiensis has an as-yet unidentified gene with similar function. C. zofingiensis seems to have critical C. reinhardtii genes for flagella motility (FLA14; ref. 33) and forming flagella (PF15, PF19), including conservation of functional residues in these two genes (34, 35). Additionally, FLA14, PF15, and PF19 were expressed in a variety of conditions, which suggests that these genes are functional despite lacking visible flagella. Furthermore, we identified putative orthologs of 25 of 40 C. reinhardtii meiosis-associated genes (30, 36), which was more than we observed for C. sp. NC64A (only 22 of 40). In C. zofingiensis, most of these genes are transcribed under many conditions, but a few such as GSP1, MER3, and DMC1 had low transcript abundance except under a low dose of Rose Bengal (5 μM Rose Bengal, 0.5 h dark and 1 h of 100 μmol photons⋅m−2⋅s−1). Eleven of the families not found in C. zofingiensis were specific to C. reinhardtii. Although these data cannot rule out the possibility that a sexual cycle was recently lost, it is more likely that the high number of apparent cilia/flagella and meiosis genes suggests the existence of sexual reproduction and a motile stage that has not yet been observed in C. zofingiensis. Life cycle studies and, in particular, investigations in search of a cryptic sexual cycle, which may require specific conditions, should be a subject of future research in C. zofingiensis.
Astaxanthin Biosynthesis Pathway and Astaxanthin-Deficient Mutants.
Astaxanthin is an important and valuable algal bioproduct. In microalgae, astaxanthin is often produced in high abundance under stressful conditions, consistent with the hypothesis that it confers protection against oxidative stress. However, astaxanthin is not coupled functionally or structurally to the photosynthetic apparatus. Instead, astaxanthin functions as an internal sunscreen and antioxidant by absorbing excess light and quenching reactive oxygen species (13, 15). Additionally, astaxanthin accumulates in cytoplasmic lipid droplets where it could prevent peroxidation of fatty acids (13, 15). Astaxanthin is synthesized via the carotenoid biosynthetic pathway, which has been reviewed (15, 37, 38); however, key steps in its biosynthesis are still undetermined. Most of what is known about astaxanthin biosynthesis in algae comes from studies of H. pluvialis, for which we lack a sequenced genome. It is thought that β-carotene is exported from the chloroplast into lipid droplets in H. pluvialis where astaxanthin is synthesized by the introduction of two keto-groups catalyzed by a di-iron beta-ketolase (BKT), which is followed by the introduction of two hydroxyl groups catalyzed by a hydroxylase (CHYB) (15, 39). However, the mechanisms of export and transport remain elusive. In contrast, it is hypothesized that, in C. zofingiensis, the hydroxylation of β-carotene occurs first and that astaxanthin is formed by the ketolation of zeaxanthin (13). In vitro enzymatic studies of C. zofingiensis show that BKT catalyzes the ketolation of β-carotene to canthaxanthin and zeaxanthin to astaxanthin, whereas CHYB catalyzes the hydroxylation of β-carotene to zeaxanthin but not of canthaxanthin to astaxanthin (13). Liu et al. (13) also concluded there was only one copy of BKT and CHYB present in C. zofingiensis; however, a recent study suggests there are two copies of BKT (40). For comparison, H. pluvialis has three BKT genes that are differentially regulated by environmental factors (41). In both microalgae, astaxanthin is esterified and stored in lipid droplets; however, the acyltransferase involved has not been identified.
The genome of C. zofingiensis provides insights into the astaxanthin pathway. The annotated carotenoid biosynthetic pathway in this alga (SI Appendix, Table S3) appears to be similar to that in C. reinhardtii (42). For example, there are four putative carotene hydroxylase genes, encoding three cytochrome P450s (two CYP97A and one CYP97C) and one di-iron type hydroxylase (CHYB). In addition, we found two putative BKT genes in the genome, in accordance with recent results (40). BKT1 and BKT2 contain highly conserved histidine motifs present in H. pluvialis and bacterial beta-ketolases (43, 44). These motifs are involved in iron binding and, in bacteria, mutations in them abolish the ability to form ketocarotenoids (44). The BKT genes from microalgae share highly conserved regions and are more similar to each other than those from bacteria. PredAlgo predicts the localization of both BKT1 and BKT2 to “other” areas of the cell, which could support localization of these enzymes to the cytosol. However, this prediction has not yet been verified experimentally. The genome also shows that there is a wide distribution of carotenoid biosynthesis genes across many chromosomes as is typical in eukaryotes.
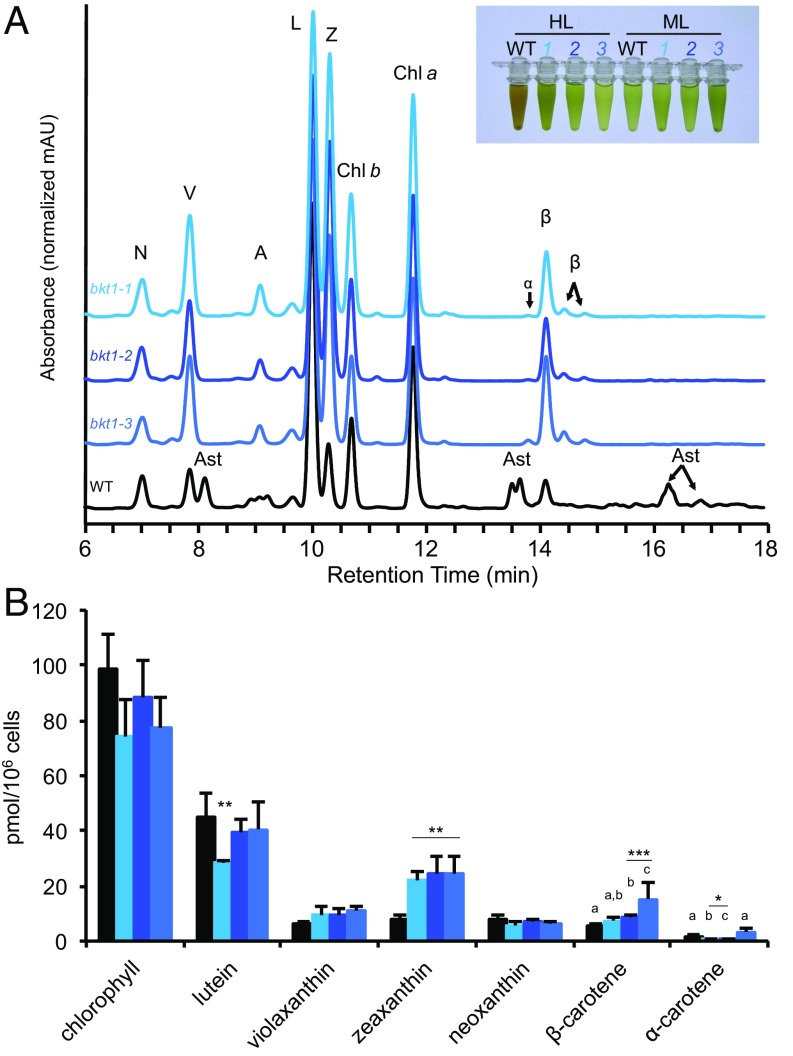
To study C. zofingiensis astaxanthin production with a nonbiased approach, a genetic screen was conducted to identify genes essential for astaxanthin synthesis. C. zofingiensis cells were randomly mutated by using UV radiation, grown on glucose medium to induce astaxanthin accumulation (13), and 65 colonies were identified that were visibly green rather than pink because of the lack of astaxanthin production, which was subsequently confirmed by HPLC analysis (SI Appendix, SI Text). Similar HPLC chromatograms were observed for all mutants. Initially, three strains were selected for sequencing of BKT1 and BKT2, and all three showed different single mutations in highly conserved areas of BKT1 but no mutations in BKT2, and, thus, these mutants were named bkt1-1, bkt1-2, and bkt1-3 (SI Appendix, Tables S4 and S5). When grown in high light, the mutants accumulated increased levels of astaxanthin precursor compounds, especially zeaxanthin but also β-carotene, and more violaxanthin, despite similar levels of other pigments (Fig. 4 and SI Appendix, SI Text). BKT1 was sequenced in an additional 13 mutants, and all showed mutations in conserved regions of the gene (SI Appendix, Table S4). These data suggest that the disruption of BKT1 alone is sufficient to abolish astaxanthin production, but we cannot unambiguously distinguish whether the committed step toward astaxanthin begins with β-carotene or zeaxanthin. Although the screen demonstrates that the BKT1 enzyme is required for astaxanthin biosynthesis, based on these results we cannot determine whether BKT2 is nonfunctional or whether it may act in a secondary reaction downstream of BKT1. Both BKT1 and BKT2 were highly expressed in response to H2O2 stress (876 and 367 FPKMs, respectively), and both were identified in the screen for H2O2 treatment-enriched genes (SI Appendix, Fig. S12). To a lesser extent, both were expressed in response to Rose Bengal treatment (394 and 35 FPKMs). It is unlikely that BKT1 and BKT2 form an obligate heterodimer because then mutations in either BKT1 or BKT2 should have been detected in different mutant strains.
Fig. 4.
C. zofingiensis astaxanthin-deficient mutants. Astaxanthin-deficient mutants were generated by using forward genetics; mutations were identified in BKT1. (A) HPLC traces of wild type, bkt1-1, bkt1-2, and bkt1-3 grown under high light (HL, 400–450 μmol photons⋅m−2⋅s−1), showing the mutants’ lack of astaxanthin production. Pigment abbreviations are as follows: A (antheraxanthin), α (α-carotene), Ast (astaxanthin), β (β-carotene), Chl a (chlorophyll a), Chl b (chlorophyll b), L (lutein), N (neoxanthin), V (violaxanthin), Z (zeaxanthin). Pigments were detected at 445 nm with reference at 550 nm (SI Appendix, SI Text). Inset shows high light WT growth with astaxanthin resulting in orange-brown color from astaxanthin (orange) and chlorophylls (green), whereas mutants bkt1-1, bkt1-2, and bkt1-3 do not produce astaxanthin and remain green. Under medium light (ML, 100 μmol photons⋅m−2⋅s−1), WT does not produce high amounts of astaxanthin and remains green with similar color as bkt1-1, bkt1-2, and bkt1-3. (B) Pigment levels (mean ± SD, n = 3 or 4) in HL-grown WT, bkt1-1, bkt1-2, and bkt1-3 showing higher levels of carotenoids with similar levels of chlorophyll. *P < 0.05, **P < 0.01, ***P < 0.001 (SI Appendix, SI Text).
High Light-Induced Gene Expression.
To investigate the physiological changes associated with acclimation to high light and to elucidate unidentified genes in the astaxanthin biosynthesis pathway in C. zofingiensis, an RNA-Seq experiment was conducted in which cultures were moved from normal growth light intensity (100 μmol photons⋅m–2⋅s–1) to high light intensity (400 μmol photons⋅m–2⋅s–1) (SI Appendix, SI Text and Dataset S21). Cultures were collected for nuclear, plastid, and mitochondrial gene expression analyses at 0, 0.5, 1, 3, 6, and 12 h (n = 4) after the shift to high light, and in control cultures, which were maintained at the normal growth light intensity (SI Appendix, SI Text).
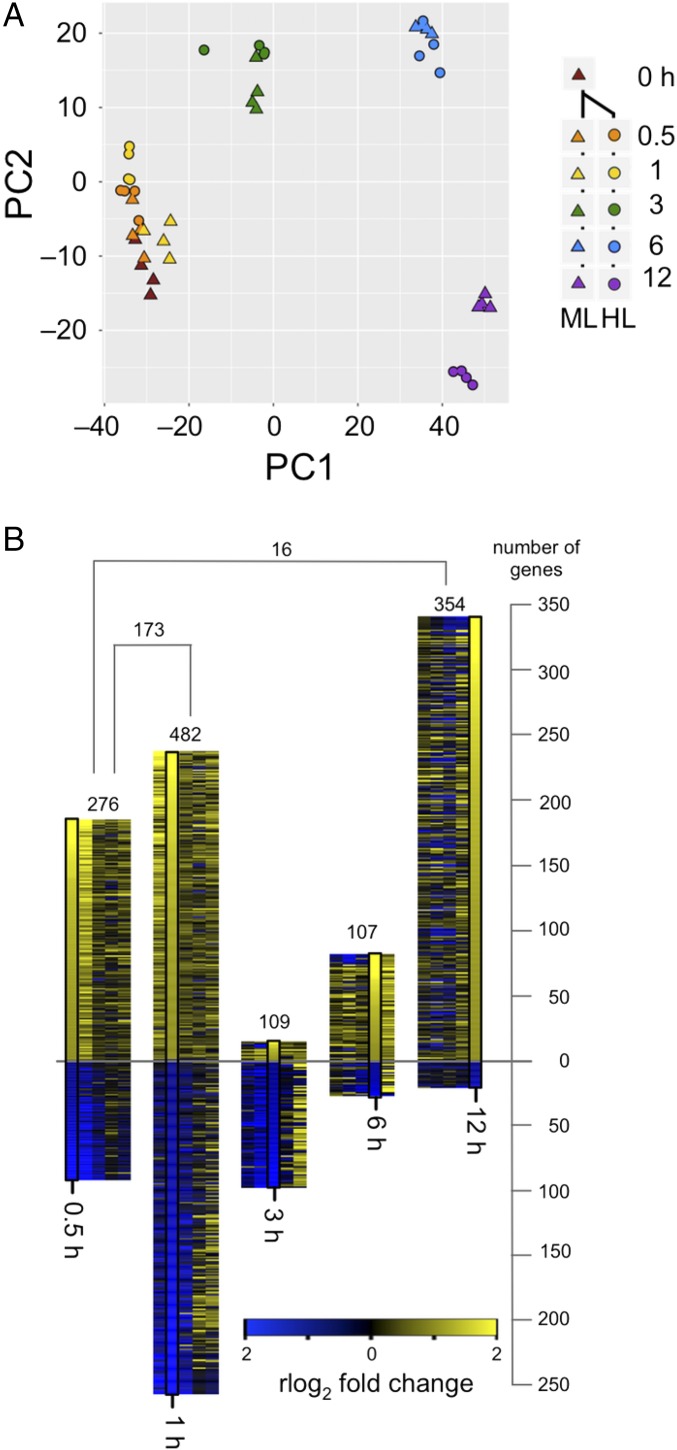
A principal component analysis of the regularized log2-transformed counts from the resulting transcriptome profiles showed that time and treatment explain nearly all observed variation in gene expression between the conditions (95%, Fig. 5A). Time induced the largest variation for both the control and treatment cultures, which may have been caused by the diurnal lighting regime; these cultures were maintained on a day-night cycle (16 h light, 8 h dark) and sampled during daylight hours. The large changes throughout the day are not surprising given that in C. reinhardtii more than 80% of the transcriptome is differentially expressed with diurnal periodicity (45). In addition to time-of-day changes, there is also a substantial effect from the shift to high light, as evidenced by the distinct groupings of control and treatment samples. Control cultures at each time point were used to separate the effects of time from high light.
Fig. 5.
C. zofingiensis RNA expression during transition to high light. Cultures of C. zofingiensis were grown diurnally (16 h light, 8 h dark) in 100 μmol photons⋅m−2⋅s−1 medium light (ML). At t = 0, cultures were transferred to 400 µmol photons⋅m−2⋅s−1 high light (HL). Samples were collected in quadruplicate at 0, 0.5, 1, 3, 6, and 12 h for ML cultures and at 0.5, 1, 3, 6, and 12 h for HL. Transcript abundances for each sample were determined by RNA-Seq. (A) Principal component analysis (PCA) of the regularized log2-transformed counts for all 44 samples. The two most significant components, accounting for 95% of variation, are shown. ML (triangles) and HL (circles) are displayed with time point indicated by color. (B) Differentially expressed genes during transition to HL. Expression fold change in HL versus ML was determined for each time point for all genes. Genes at least twofold up-regulated or twofold down-regulated are indicated by the height of the bar above or below the line, respectively (P < 0.01); the total is indicated above each bar. The regularized log2-transformed fold change between HL and ML is shown in the black box for each time point as indicated by color. For comparison, the fold change of each of these genes at the other time points is presented flanking the black box. The number of differentially expressed genes in common between 0.5 and 1 h and between 0.5 and 12 h is indicated by square brackets.
To further evaluate the effect of high light, differentially expressed genes at each time point were identified. Those genes whose expression had a greater than twofold change (P < 0.01) in either direction between the high light-treated cultures and controls were determined and visualized in a heat map, scaled relative to the number of genes in each group (Fig. 5B). Most genes were differentially expressed either early in the experiment (276 genes at 0.5 h and 492 genes at 1 h) or late (362 genes at 12 h). The greatest overlap of significantly differentially expressed genes was during the early time points (0.5 and 1 h), unsurprising given that these samples were collected closest together in time. Additionally, during these early time points, high light had a greater effect than time (Fig. 5A). Over the course of the experiment, there was greater up-regulation of significantly differentially expressed genes with 67%, 75%, and 94% of genes up-regulated at 0.5, 6, and 12 h, respectively, but more genes were significantly down-regulated at 1 h (52%) and 3 h (86%). Most genes had relatively modest changes (<fourfold) in the cultures shifted to high light, although expression of ELIP8 (early light-induced protein) and ELIP10 had >20-fold increases at 0.5 h. Among chloroplast-encoded genes, both psaA and atpF had significant up-regulation at 1 h. No significantly differentially expressed mitochondrial genes were found during the shift to high light.
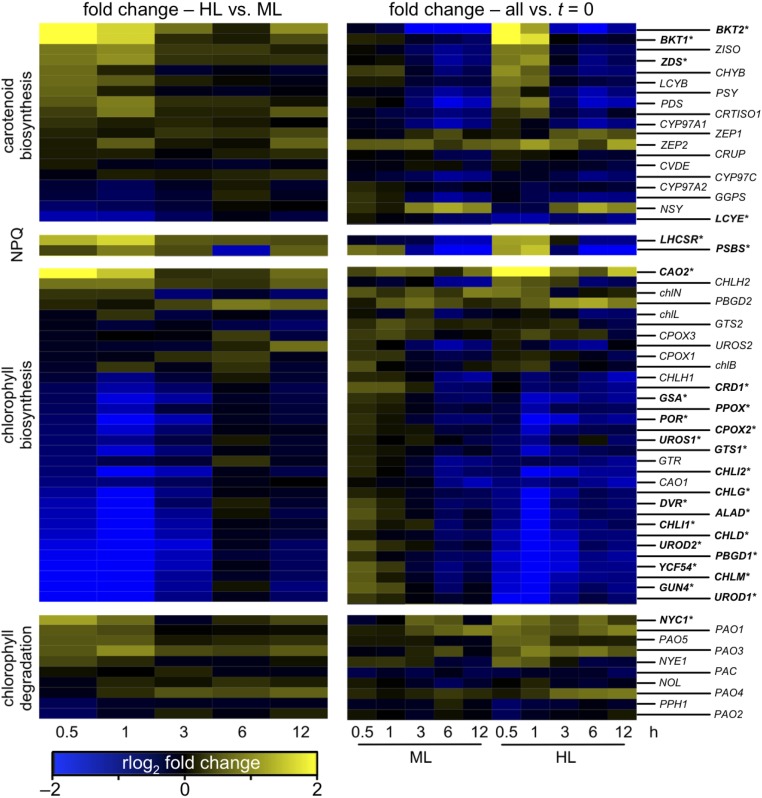
Because of the high level of interest in astaxanthin production in C. zofingiensis, we examined the genes involved in carotenoid biosynthesis during the shift to high light. High light causes an accumulation of secondary carotenoids (in particular, astaxanthin) in C. zofingiensis (46–48). In the present study, both BKT1 and BKT2 have the highest increase in gene expression, which occurs immediately after the light shift at 0.5 h (Fig. 6). Despite the increase in BKT2 gene expression in high light, its role in carotenoid biosynthesis has not been established. Many genes at various points in the carotenoid biosynthesis pathway were up-regulated early (0.5 h and 1 h) in response to the high light treatment, including phytoene synthase (PSY) (Fig. 6), which catalyzes the committed step in carotenoid biosynthesis. Previous studies have reported similar up-regulation of PSY, PDS, BKT, and CHYB at longer time points in response to increases in light (46, 49, 50). However, our study also revealed a significant increase in expression of ZDS at 1 h and a significant decrease in LCYE shortly after the shift to high light. Additionally, down-regulation of many genes in the carotenoid biosynthesis pathway was observed at later time points (6 and 12 h) in both the treatment and control cultures; this regulation is likely an effect of the diurnal cycle. A higher expression of carotenoid biosynthesis genes would support an increase in secondary carotenoids, but does not exclude the possibility that posttranslational modifications of carotenoid biosynthetic enzymes may also account for the accumulation of secondary carotenoids during high light.
Fig. 6.
C. zofingiensis RNA-Seq expression of select genes during the transition to high light. RNA-Seq was performed on cultures following a shift from medium light (ML, 100 μmol photons⋅m−2⋅s−1) to high light (HL, 400–450 μmol photons⋅m−2⋅s−1) as described in Fig. 5. C. zofingiensis genes potentially involved in carotenoid biosynthesis, nonphotochemical quenching (NPQ), and chlorophyll biosynthesis and degradation were identified by manual curation. (Left) The regularized log2-transformed fold change between HL and ML for each of these genes at each time point is plotted as a heatmap. (Right) The regularized log2-transformed fold change between each time point relative to t = 0 is plotted. Significantly differential genes that are over twofold up- or down-regulated are indicated by an asterisk and bold text (P < 0.01).
The high-quality genome and transcriptome we generated in combination with the high light RNA-Seq experiment allowed us to identify candidates for additional genes involved in astaxanthin biosynthesis and accumulation. As mentioned above, little is known about the mechanism of translocation of the astaxanthin precursor(s) out of the chloroplast, the hydroxylation of the astaxanthin precursor, transport of astaxanthin into lipid droplets, or the esterification of astaxanthin. We identified genes putatively involved in astaxanthin biosynthesis by examining significantly differentially expressed genes with high increases in gene expression during the shift to high light, and looking for annotated activity compatible with hypothetical mechanisms of astaxanthin biosynthesis. Genes that are up-regulated early during high light that may be implicated in the astaxanthin pathway include four ABC transporters (Cz04g21110, Cz05g17060, Cz09g27180, and Cz08g16130), two cytochrome P450 proteins (Cz10g28330 and Cz11g14160), and an acyltransferase (Cz02g29020). The ABC transporters may form a complex that exports the astaxanthin precursor(s) from the chloroplast. The cytochrome P450 proteins could be involved in hydroxylation of astaxanthin precursors in the cytosol, and the acyltransferase could be involved in esterification of astaxanthin. However, these genes need to be experimentally tested in vitro and/or in vivo to determine whether they function in the pathway. Improving understanding of the astaxanthin pathway may provide opportunities to enhance astaxanthin production in this species and others.
In addition to changes in carotenoid biosynthesis, we also investigated other algal high light responses, including photoprotective mechanisms and chlorophyll metabolism. In photosynthetic organisms, excess light must be safely dissipated to prevent oxidative damage. C. reinhardtii transiently expresses PSBS at the onset of high light and LHCSR proteins accumulate under high light, and this accumulation is correlated with nonphotochemical quenching capacity (51, 52). Whereas C. reinhardtii has multiple copies of both LHCSR and PSBS (51, 52), we found only single copies of LHCSR and PSBS in C. zofingiensis, despite having high nonphotochemical quenching capacity (9). As expected, both LHCSR and PSBS were up-regulated at the early time points during the shift to high light and, in particular, at 1 h, which is consistent with observations of C. reinhardtii during the dark-to-light transition (45). Similar to the carotenoid biosynthesis genes, under the diurnal cycle LHCSR and PSBS are down-regulated by the end of the day (6 and 12 h) in both conditions (Fig. 6). Reduction in chlorophyll is another common physiological response of algae exposed to high light (53). Accordingly, during the shift to high light, many C. zofingiensis genes involved in chlorophyll synthesis were down-regulated and chlorophyll degradation genes were up-regulated (Fig. 6). The combination of these changes would lead to a reduction in chlorophyll content either during acclimation or as a stress response to high light.
Annotation of Metabolic Pathways and Photosynthesis-Related Genes.
Genes encoding homologs of the primary metabolic pathway enzymes involved with carbon, carotenoids, chlorophyll, fatty acids, lipids, and proteins involved in the composition, assembly, and regulation of the photosynthetic apparatus, were preliminarily identified by using the BLAT sequence search tool (54) against a C. zofingiensis draft genome (SI Appendix, Table S3). Based on the quality of alignments and comparison with well-characterized, closely-related plant and algal query sequences, the targeted gene models in C. zofingiensis were submitted as queries in reciprocal BLAST searches against the NCBI RefSeq nonredundant protein database to confirm coverage, domain architecture, and similarity across closely-related homologs. Because of the high quality of the C. zofingiensis assembly, this procedure resulted in a nearly complete list of putative genes needed to complete each pathway. Identified gene models were used to assess the quality of the automated gene family analysis across the six species of SI Appendix, Table S1, and the automated analysis was used to confirm additional candidate models and expand the set of annotations. Based on high sequence similarities and conservation of functional domains, we are generally confident in assignments of homology. However, it is possible that additional functional isoforms composed of more divergent sequences may also be present, having been missed by the parameters used for BLAT, BLAST, and the automated gene family analysis.
Identification and annotation of genes involved in lipid biosynthesis can provide targets for exploitation of C. zofingiensis for biofuel production. Using other oleaginous organisms as a guide, we would expect a robust oil-producing microalga to have an expanded family of acyltransferases. The C. zofingiensis diacylglycerol acyltransferases (DGAT) are too divergent from the protein sequences of type 1 DGAT and DGTT (type 2 DGATs; ref. 55) in C. reinhardtii, A. thaliana, and M. neglectum to identify the corresponding genes via BLAT. Using a more sensitive BLAST search with both types of DGAT sequences from C. subellipsoidea C-169 (gi|545360296), Chlorella vulgaris (gb|ALP13863.1), Nannochloropsis gaditana (gb|EWM23187.1), A. thaliana (gi|15224779, gi|18409359), and C. reinhardtii (Cre01.g045903, Cre03.g205050), additional copies of DGAT type 1- and DGTT-encoding genes were identified in C. zofingiensis, and yet more were identified by using the automated gene family analysis. In total, 11 genes were identified that have either an LPLAT (lysophospholipid acyltransferase) domain or a closely-related MBOAT (membrane bound O-acyltransferase) domain (SI Appendix, Table S3). We have tentatively assigned these genes as encoding proteins with diacylglycerol acyltransferase activity; however, some of these C. zofingiensis gene models have higher similarity to predicted proteins of unknown function than to annotated type 1 or type 2 DGAT proteins from other closely related organisms. Our finding of multiple copies of putative DGAT and DGTT genes in C. zofingiensis is consistent with transcriptome results from the closely-related ATCC 30412 strain (40). It is also possible, although unlikely, that one or multiple additional copies of DGAT or DGTT may be yet unidentified because of a gene modeling or assembly problem.
Homologous gene models were also identified for components of the photosynthetic apparatus and its assembly, including proteins that compose PSI, PSII, the major and minor light harvesting antennae, the cytochrome b6f complex, the chloroplast ATP synthase complex, soluble electron carriers, and known assembly factors for these complexes (SI Appendix, Table S3). Thirteen of the C. zofingiensis light-harvesting complex (LHC) genes are predicted to be more like PSI-associated LHC genes (LHCAs) while nine are predicted to be PSII-associated LHC genes (LHCBs) (SI Appendix, Table S3). This distribution is in contrast to that found in C. reinhardtii, which has nine of each of LHCA and LHCB. Further experimental work is needed to confirm the expression profiles and photosystem association of each of these putative LHC proteins, especially under different light and stress conditions. Of note is one LHC model (Lhcb-like3, Cz04g24050) with little to no transcriptional expression detected in any of our RNA-Seq conditions.
Conclusions
Our analyses of the C. zofingiensis genome, transcriptome, astaxanthin-deficient mutants, and RNA expression changes under high light reveal insights into the basic biology of the green lineage of photosynthetic organisms and the carotenoid biosynthesis pathway. We present, in the tradition of model organisms, a high-quality chromosome-level assembly with independent genome validation. The compact ∼58-Mbp genome has balanced G+C content and is rich in protein-coding sequence with few long exons per gene and relatively little repetitive sequence. We identified ortholog families for the majority of C. zofingiensis genes. The gene density is uniform over chromosomes and a syntenic comparison with other algae uncovered highly significant genomically localized blocks of genes in putative orthologous relationships; however, gene order and strands within blocks are scrambled. We have shown that BKT1 is critical for the production of astaxanthin and have identified candidate genes that could be missing components in astaxanthin biosynthesis and accumulation pathways. The addition of genomics to the experimental toolkit for C. zofingiensis makes it an attractive alga not only for fundamental studies of its biology but also the economically viable and environmentally sustainable production of biofuels and important bioproducts.
Supplementary Material
Acknowledgments
The cryo-soft X-ray tomography was supported by the US Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under field work proposal SISGRKN. The whole-genome optical mapping and high light RNA sequencing was supported by the US Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under field work proposal 449B. The transcriptome sequencing and long read sequencing was supported by Agriculture and Food Research Initiative Competitive Grant 2013-67012-21272 from the US Department of Agriculture National Institute of Food and Agriculture (to M.S.R.). The National Center for X-ray Tomography is supported by the National Institute of General Medical Sciences of the National Institutes of Health Grant P41GM103445 and the US Department of Energy, Office of Biological and Environmental Research Grant DE-AC02-05CH11231. S.J.C., S.D.G., S.S.M., and M.P. were supported by a cooperative agreement with the US Department of Energy Office of Science, Office of Biological and Environmental Research program under Award DE-FC02-02ER63421. D.L. was supported by a National Institutes of Health T32 Training Fellowship in Genome Analysis 5T32HG002536–13, the Eugene V. Cota-Robles Fellowship, and the Fred Eiserling and Judith Lengyel Doctoral Fellowship. D.W. was supported by a National Science Foundation Graduate Research Fellowship. K.K.N. is an Investigator of the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation through Grant GBMF3070.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Chromochloris zofingiensis version 5 assembly and associated annotations and gene families are included as Datasets S1–S19 (see SI Appendix, Datasets Key), and are also available on Phytozome and the project webpage at genomes.mcdb.ucla.edu/Chromochloris/. Raw Illumina and Pacific Biosciences genomic reads are available at NCBI Sequence Read Archive (accession nos. SRR5310949–SRR5310954), and raw RNA-Seq reads are under Gene Expression Omnibus accession no. GSE92515, with FPKM matrices also available as Datasets S20 and S21.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619928114/-/DCSupplemental.
References
- 1.Stephens E, et al. Future prospects of microalgal biofuel production systems. Trends Plant Sci. 2010;15:554–564. doi: 10.1016/j.tplants.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Wijffels RH, Barbosa MJ. An outlook on microalgal biofuels. Science. 2010;329:796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- 3.Breuer G, Lamers PP, Martens DE, Draaisma RB, Wijffels RH. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour Technol. 2012;124:217–226. doi: 10.1016/j.biortech.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Mao X, Zhou W, Guarnieri MT. Simultaneous production of triacylglycerol and high-value carotenoids by the astaxanthin-producing oleaginous green microalga Chlorella zofingiensis. Bioresour Technol. 2016;214:319–327. doi: 10.1016/j.biortech.2016.04.112. [DOI] [PubMed] [Google Scholar]
- 5.Mulders KJM, et al. Effect of biomass concentration on secondary carotenoids and triacylglycerol (TAG) accumulation in nitrogen-depleted Chlorella zofingiensis. Algal Res. 2014;6:8–16. [Google Scholar]
- 6.Dönz OC. Chlorella zofingiensis, eine neue Bodenalge. Ber Schweiz Bot Ges. 1934;43:127–131. [Google Scholar]
- 7.Fučíková K, Lewis LA. Intersection of Chlorella, Muriella and Bracteacoccus: Resurrecting the genus Chromochloris Kol & Chodat (Chlorophyceae, Chlorophyta) Fottea. 2012;12:83–93. [Google Scholar]
- 8.Goodenough UW. Chloroplast division and pyrenoid formation in Chlamydomonas reinhardii. J Phycol. 1970;6:1–6. [Google Scholar]
- 9.Bonente G, et al. The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem Photobiol. 2008;84:1359–1370. doi: 10.1111/j.1751-1097.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 10.Ip PF, Wong KH, Chen F. Enhanced production of astaxanthin by the green microalga Chlorella zofingiensis in mixotrophic culture. Process Biochem. 2004;39:1761–1766. [Google Scholar]
- 11.Hussein G, Sankawa U, Goto H, Matsumoto K, Watanabe H. Astaxanthin, a carotenoid with potential in human health and nutrition. J Nat Prod. 2006;69:443–449. doi: 10.1021/np050354+. [DOI] [PubMed] [Google Scholar]
- 12.Yuan J-P, Peng J, Yin K, Wang J-H. Potential health-promoting effects of astaxanthin: A high-value carotenoid mostly from microalgae. Mol Nutr Food Res. 2011;55:150–165. doi: 10.1002/mnfr.201000414. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, et al. Chlorella zofingiensis as an alternative microalgal producer of astaxanthin: Biology and industrial potential. Mar Drugs. 2014;12:3487–3515. doi: 10.3390/md12063487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capelli B, Bagchi D, Cysewski GR. Synthetic astaxanthin is significantly inferior to algal-based astaxanthin as an antioxidant and may not be suitable as a human nutraceutical supplement. Nutrafoods. 2013;12:145–152. [Google Scholar]
- 15.Solovchenko AE. Recent breakthroughs in the biology of astaxanthin accumulation by microalgal cell. Photosynth Res. 2015;125:437–449. doi: 10.1007/s11120-015-0156-3. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, et al. Utilization of cane molasses towards cost-saving astaxanthin production by a Chlorella zofingiensis mutant. J Appl Phycol. 2013;25:1447–1456. [Google Scholar]
- 17.Yuan Z, et al. Scale-up potential of cultivating Chlorella zofingiensis in piggery wastewater for biodiesel production. Bioresour Technol. 2013;137:318–325. doi: 10.1016/j.biortech.2013.03.144. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, et al. Genetic engineering of the green alga Chlorella zofingiensis: A modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker. Appl Microbiol Biotechnol. 2014;98:5069–5079. doi: 10.1007/s00253-014-5593-y. [DOI] [PubMed] [Google Scholar]
- 19.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 21.Fučíková K, Lewis PO, González-Halphen D, Lewis LA. Gene arrangement convergence, diverse intron content, and genetic code modifications in mitochondrial genomes of sphaeropleales (chlorophyta) Genome Biol Evol. 2014;6:2170–2180. doi: 10.1093/gbe/evu172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fučíková K, Lewis PO, Lewis LA. Chloroplast phylogenomic data from the green algal order Sphaeropleales (Chlorophyceae, Chlorophyta) reveal complex patterns of sequence evolution. Mol Phylogenet Evol. 2016;98:176–183. doi: 10.1016/j.ympev.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 23.Blanc G, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 27.Leliaert F, et al. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31:1–46. [Google Scholar]
- 28.Tardif M, et al. PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol. 2012;29:3625–3639. doi: 10.1093/molbev/mss178. [DOI] [PubMed] [Google Scholar]
- 29.Lopez D, et al. Dynamic changes in the transcriptome and methylome of Chlamydomonas reinhardtii throughout its life cycle. Plant Physiol. 2015;169:2730–2743. doi: 10.1104/pp.15.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanc G, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel BD, et al. Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 2009;93:157–177. doi: 10.1016/S0091-679X(08)93009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dymek EE, Smith EF. PF19 encodes the p60 catalytic subunit of katanin and is required for assembly of the flagellar central apparatus in Chlamydomonas. J Cell Sci. 2012;125:3357–3366. doi: 10.1242/jcs.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dymek EE, Lefebvre PA, Smith EF. PF15p is the chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryot Cell. 2004;3:870–879. doi: 10.1128/EC.3.4.870-879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferris PJ, Armbrust EV, Goodenough UW. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics. 2002;160:181–200. doi: 10.1093/genetics/160.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemoine Y, Schoefs B. Secondary ketocarotenoid astaxanthin biosynthesis in algae: A multifunctional response to stress. Photosynth Res. 2010;106:155–177. doi: 10.1007/s11120-010-9583-3. [DOI] [PubMed] [Google Scholar]
- 38.Takaichi S. Carotenoids in algae: Distributions, biosyntheses and functions. Mar Drugs. 2011;9:1101–1118. doi: 10.3390/md9061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grunewald K, Hagen C. β-carotene is the intermediate exported from the chloroplast during accumulation of secondary carotenoids in Haematococcus pluvialis. J Appl Phycol. 2001;13:89–93. [Google Scholar]
- 40.Huang W, et al. Transcriptome analysis of Chlorella zofingiensis to identify genes and their expressions involved in astaxanthin and triacylglycerol biosynthesis. Algal Res. 2016;17:236–243. [Google Scholar]
- 41.Huang JC, Chen F, Sandmann G. Stress-related differential expression of multiple beta-carotene ketolase genes in the unicellular green alga Haematococcus pluvialis. J Biotechnol. 2006;122:176–185. doi: 10.1016/j.jbiotec.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Lohr M, Im CS, Grossman AR. Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii. Plant Physiol. 2005;138:490–515. doi: 10.1104/pp.104.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, Zhong Y, Sandmann G, Liu J, Chen F. Cloning and selection of carotenoid ketolase genes for the engineering of high-yield astaxanthin in plants. Planta. 2012;236:691–699. doi: 10.1007/s00425-012-1654-6. [DOI] [PubMed] [Google Scholar]
- 44.Ye RW, Stead KJ, Yao H, He H. Mutational and functional analysis of the beta-carotene ketolase involved in the production of canthaxanthin and astaxanthin. Appl Environ Microbiol. 2006;72:5829–5837. doi: 10.1128/AEM.00918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zones JM, Blaby IK, Merchant SS, Umen JG. High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell. 2015;27:2743–2769. doi: 10.1105/tpc.15.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Huang J, Sandmann G, Chen F. High-light and sodium chloride stress differentially regulate the biosynthesis of astaxanthin in Chlorella zofingiensis (Chlorophyceae) J Phycol. 2009;45:635–641. doi: 10.1111/j.1529-8817.2009.00689.x. [DOI] [PubMed] [Google Scholar]
- 47.Del Campo JA, et al. Accumulation of astaxanthin and lutein in Chlorella zofingiensis (Chlorophyta) Appl Microbiol Biotechnol. 2004;64:848–854. doi: 10.1007/s00253-003-1510-5. [DOI] [PubMed] [Google Scholar]
- 48.Rise M, et al. Accumulation of secondary carotenoids in Chlorella zofingiensis. J Plant Physiol. 1994;144:287–292. [Google Scholar]
- 49.Cordero BF, Couso I, León R, Rodríguez H, Vargas MA. Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl Microbiol Biotechnol. 2011;91:341–351. doi: 10.1007/s00253-011-3262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J, Liu J, Li Y, Chen F. Isolation and characterization of the phytoene desaturase gene as a potential selective marker for genetic engineering of the astaxanthin-producing green alga Chlorella zofingiensis (Chlorophyta) J Phycol. 2008;44:684–690. doi: 10.1111/j.1529-8817.2008.00511.x. [DOI] [PubMed] [Google Scholar]
- 51.Correa-Galvis V, et al. Photosystem II subunit PsbS is involved in the induction of LHCSR-dependent energy dissipation in Chlamydomonas reinhardtii. J Biol Chem. 2016;291:17478–17487. doi: 10.1074/jbc.M116.737312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 53.Erickson E, Wakao S, Niyogi KK. Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J. 2015;82:449–465. doi: 10.1111/tpj.12825. [DOI] [PubMed] [Google Scholar]
- 54.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyle NR, et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem. 2012;287:15811–15825. doi: 10.1074/jbc.M111.334052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.