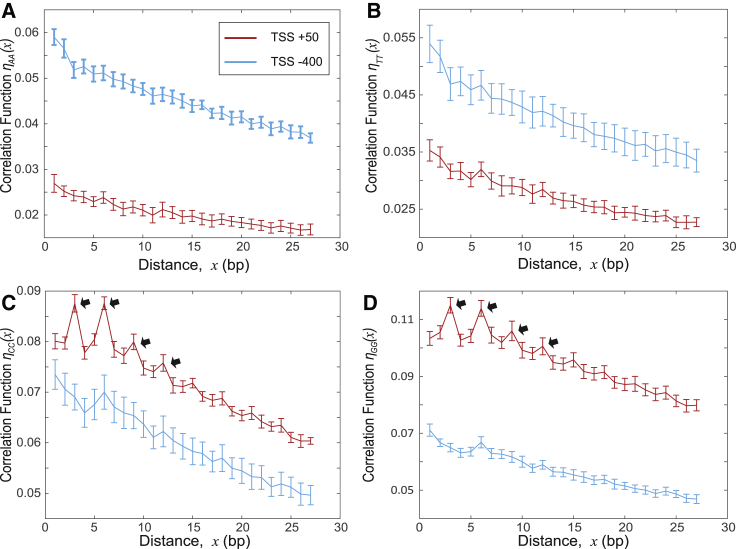
Figure 3.
Normalized pair (binary) correlation functions for the nucleotide spatial distribution. (A–D) Shown here is the computed correlation function ηαα(x) = (Nαα(x) − < Nαα(x) > rand)/L0, where Nαα(x) represents the average number of nucleotide pairs of type α separated by the relative distance x bp, and L0 is the width of the window. We used L0 = 100 bp. We used DNA sequences of 6097 genes for two genomic regions: the region of high TFIIB binding intensity (0, 100 bp) (red lines); and the region of low TFIIB binding intensity (−450, −350 bp) (blue lines). To compute error bars, we calculated the mean for each chromosome and divided the results into five randomly chosen subgroups and computed the mean for each subgroup. The error bars are defined as 1 SD of the mean between the subgroups. The arrows in (C) and (D) emphasize the peaks of the correlation function. These peaks represent the enrichment of repeated DNA triplets. To see this figure in color, go online.