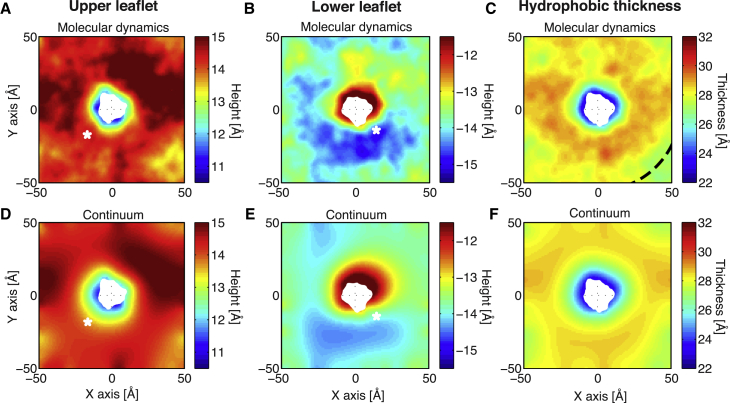
Figure 2.
Comparison of protein-induced membrane deformations from molecular dynamics and continuum elasticity. (A–C) Shown here are the average membrane height profiles for the upper leaflet (A), lower leaflet (B), and hydrophobic thickness (C) from one 250-ns MD simulation of gramicidin in a POPC bilayer. (D)–(F) show the corresponding continuum membrane surfaces. For the continuum calculations (D)–(F), we used boundary conditions at the protein-membrane boundary and at the edges of the box that were extracted from the MD data in (A)–(C). The region where the membrane is compressed near the protein (compression ring) can be clearly seen in the hydrophobic mismatch panels of both the MD (C) and continuum calculations (F). Dashed arc line in (C) shows a 55 Å radial distance measured from the center of the protein. White asterisks show the location of maximum difference between leaflet heights calculated using simulation (A) and (B) when compared to continuum (D) and (E). For reference, in Fig. S8 we show the gramicidin-induced membrane deformations calculated using our hybrid-atomistic model without any input from MD. To see this figure in color, go online.