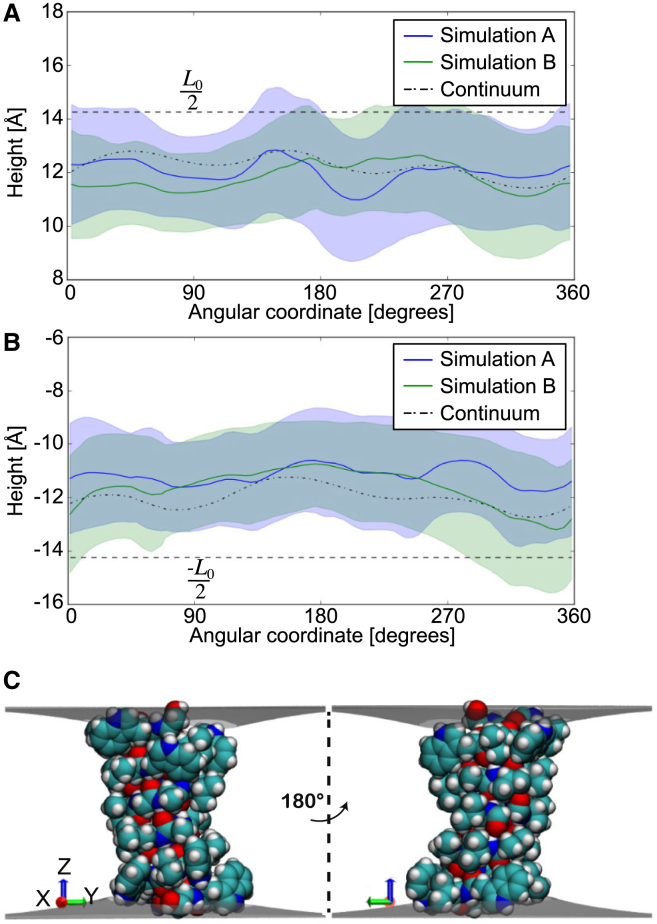
Figure 4.
Comparison of protein-membrane boundary distortions from the continuum model and MD simulation. Given here are the membrane height values at the protein-membrane boundary for the upper leaflet (A) and lower leaflet (B). The dotted line corresponds to the average solution calculated from 32 protein snapshots by our continuum model. The blue and green solid lines are the average membrane height values from the two independent 250 ns MD simulations. The blue- and green-shaded regions correspond to MD solutions within 1 SD and the overlap between the blue- and green-shaded region has been highlighted in a darker color. The equilibrium height of the undeformed monolayer L0/2 is shown by dashed lines. (C) Shown here are the membrane surface profiles calculated from the model. The angular coordinate in (A) and (B) are with respect to the x axis shown here. The protein-membrane boundaries align with the indole nitrogens of the tryptophans and the hydroxyl of the terminal ethanolamine. The right view corresponds to a 180° rotation about the z axis. To see this figure in color, go online.