Abstract
Maternal uniparental disomy of chromosome 14 (upd(14)mat) or Temple syndrome is an imprinting disorder associated with a relatively mild phenotype. The absence of specific congenital malformations makes this condition underdiagnosed in clinical practice. A boy with a de novo robertsonian translocation 45,XY,rob(13;14)(q10;q10) is reported; a CGH/SNP array showed a loss of heterozygosity in 14q11.2q13.1. The final diagnosis of upd(14)mat was made by microsatellite analysis, which showed a combination of heterodisomy and isodisomy for different regions of chromosome 14. Obesity after initial failure to thrive developed, while compulsive eating habits were not present, which was helpful for the clinical differential diagnosis of Prader-Willi syndrome. In addition, the boy presented with many phenotypic features associated with upd(14)mat along with hypoesthesia to pain, previously unreported in this disorder, and bilateral cryptorchidism, also rarely described. These features, as well as other clinical manifestations (i.e., truncal obesity, altered pubertal timing), may suggest a hypothalamic-pituitary involvement. A detailed cytogenetic and molecular characterization of the genomic rearrangement is presented. Early genetic diagnosis permits a specific follow-up of children with upd(14)mat in order to optimize the long-term outcome.
Keywords: Array CGH, Genetic obesity, Growth, Heterodisomy, Intellectual disability, Isodisomy, Maternal uniparental disomy 14
Uniparental disomy (UPD) is the condition in which both homologous chromosomes are derived from only one parent [Robinson, 2000]. UPD can be classified as either isodisomy or heterodisomy according to the homozygosity or heterozygosity of polymorphic alleles inherited from the parent; it can involve the whole chromosome or some parts of the chromosome (segmental UPD) [Engel, 1980]. Evaluation of the parental origin of UPD is important to better assess the risk for health adverse effects of UPD [Robinson, 2000].
The human chromosome 14 carries a 1-Mb cluster of imprinted genes located in 14q32 [Hoffman and Heller, 2011]. This cluster includes paternally expressed genes such as DLK1 (delta-like non-canonical Notch ligand 1), RTL1 (retrotransposon-like 1), and DIO3 (Deiodinase, iodothyronine, type III) as well as maternally expressed noncoding RNAs such as MEG3 (maternally expressed 3), RTL1as (RTL1 antisense), MEG8 (maternally expressed 8), and numerous C/D box small nucleolar (sno)RNAs and microRNAs. Indeed, it remains to be clarified whether DIO3 is truly a paternally expressed gene. The parental expression of imprinted genes is determined by 2 differentially methylated regions [Buiting et al., 2008; Hoffman and Heller, 2011; Kagami et al., 2012]. These regions are methylated on the paternal chromosome and unmethylated on the maternal one [Temple et al., 2007; Beygo et al., 2015].
The maternal and paternal UPDs for chromosome 14 cause distinct phenotypes: upd(14)mat (Temple syndrome; EUCID.net; www.imprinting-disorders.eu) is characterized by pre- and postnatal growth retardation, developmental delay, muscular hypotonia, joint laxity, small hands and feet, truncal obesity, precocious or early onset of puberty, and adult short stature [Ioannides et al., 2014]. On the other hand, upd(14)pat (Kagami-Ogata syndrome; EUCID.net) causes a more serious phenotype with polyhydramnios, thoracic dysplasia (coat hanger sign) with respiratory failure, abdominal defects, growth retardation, developmental delay, and facial abnormalities with full cheeks and protruding philtrum [Ogata and Kagami, 2016].
Here, we report on a boy with a de novo robertsonian translocation involving chromosomes 13 and 14; he also presented with upd(14)mat. His phenotype is discussed in relation to the previously reported individuals with upd(14)mat. A detailed cytogenetic and molecular characterization of the genomic rearrangement is described.
Patient and Methods
Clinical Report
The boy was conceived by assisted reproductive technology due to fertility problems in the nonconsanguineous 39-year-old parents. At 31 weeks of gestation, oligodramnios and intrauterine growth retardation were diagnosed. He was born at 40 weeks of gestation with both low birth length and weight (−2.05 SD and −2.26 SD, respectively) according to Italian neonatal standards [Bertino et al., 2010]. At birth, he showed a prominent metopic suture, but brain ultrasonography did not show malformations.
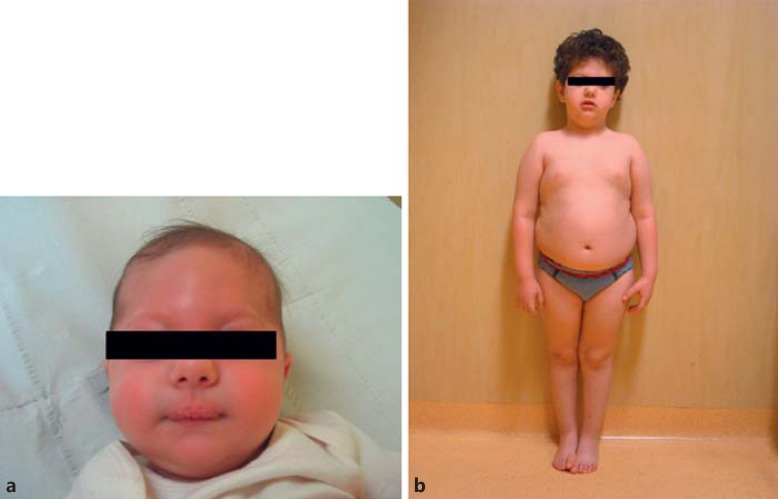
The boy came to our attention at 2 months of age (Fig. 1a): his length (54.5 cm) was −2.02 SD, weight (4,450 g) was −1.78 SD for age and −0.19 SD for length; his head circumference (38 cm) was within normal range, −0.97 SD (WHO standards; www.who.int/childgrowth/software/eu/). He also showed trigonocephaly, dysmorphic facial characteristics – such as an asymmetrical face, epicantus, hypotelorism, a mild strabism, upslanting palpebral fissures, and retrognathia – and a flat angioma on the forehead. Both gonads were palpable in the inguinal region. At 6 months, heart ultrasonography as well as ophthalmologic and otorhinolaryngoiatric examinations were normal; his routine blood and urine analyses were in normal range, except for the values of the seric IgA [12 mg/dL (normal range: 82–453 mg/dL)], IgG [334 mg/dL (normal range: 751–1560 mg/dL)], and IgM [31 mg/dL (normal range: 46–304 mg/dL)]. Abdominal ultrasonography was normal. At 9 months, the neurological evaluation showed hypoesthesia to pain. The boy had a good environment interaction and adequately manipulated his toys, but his motor and developmental milestones were delayed: he was neither able to sit nor hold his head up; he walked without support at the age of 21 months and uttered a few words at 24 months. At 6, 9, and 24 months, linear growth progressively improved, while weight was below the normal values for height and age (Table 1). Coeliac disease was ruled out by appropriate serum analyses. The boy's head circumference also improved (6 months: −2.78 SD, 9 months: −2.35 SD, and 24 months: −1.14 SD; WHO standards). At the age of 3 years, his height normalized, while weight increased to the upper values for both height and age (Table 1); his head circumference also normalized (−0.72 SD). Ultrasonography confirmed the presence of both testes located in the inguinal canals.
Fig. 1.
Our proband with upd(14)mat at the age of 2 months (a) and at the age of 5.7 years (b).
Table 1.
Proband with upd(14)mat: evolution of height and weight during the first 5 years of life
| Age, years | Height |
Weight |
|||
|---|---|---|---|---|---|
| cm | SDa | kg | SD for lengtha | SD for agea | |
| 0.2 | 54.5 | −2.02 | 4.4 | −0.19 | −1.78 |
| 0.5 | 64.0 | −0.70 | 5.8 | −2.36 | −2.78 |
| 0.8 | 71.0 | −0.43 | 6.9 | −2.91 | −2.35 |
| 2.0 | 86.5 | −0.44 | 10.0 | −2.17 | −1.71 |
| 3.0 | 97.5 | 17.4 | |||
| 5.7 | 110.0 | −0.82b | 25 | 3.56b | 2.24b |
upd, uniparental disomy.
Calculated according to WHO standards.
Calculated according to Tanner and Whitehouse [1976].
At 5.7 years of age, the boy showed normal height, but he developed overt truncal obesity (Fig. 1b; Table 1). His facial features included a broad and high forehead as well as a short nose with a wide nasal tip. He clinically presented with genu valgum and small hands and feet, but raw measures were not performed; his neuropsychological profile revealed mild intellectual disability (total IQ 57). Thus, he started primary school with the help of a support teacher.
Methods
Karyotype analysis was done by using standard methods. A CGH/SNP array using a 180 K platform (Agilent Technologies, Santa Clara, CA, USA) was performed. Briefly, 750 ng of DNA from the patient and from a normal male control (Agilent) was digested with RSAI and ALUI restriction enzymes. Test and reference DNA were differentially labeled with Cy5-dCTP or with Cy3-dCTP using random primer labeling according to the manufacturer's protocol (Agilent). The labeling reactions were applied to the array and incubated for 24 h at 65°C. Finally, the slides were washed and scanned using the Agilent scanner. The identification of individual spots on scanned arrays was performed with the dedicated software (Cytogenomics Software, Agilent) as well as filtration, normalization and exclusion of spots with aberrant morphology or high background. The proband and his parents were genotyped for 28 short tandem repeats (STRs) spanning the entire long arm of chromosome 14 by polymerase chain reaction amplification and separation on an automated ABI-3130 DNA sequencer. The polymorphic markers were analyzed by GeneScan3.1 software (Applied Biosystems, Foster City, CA, USA). The location of the STRs was obtained from UCSC Genome Bioinformatics (https://genome-euro.ucsc.edu; build 37/hg19).
Results
Karyotype analysis detected a robertsonian translocation 45,XY,rob(13;14)(q10;q10) in all of the 100 analyzed metaphases. Both parents had a normal karyotype.
The CGH/SNP array did not reveal any deletion or duplication, but it showed a loss of heterozygosity (LOH) of about 13.6 Mb in the distal portion of chromosome 14: 14q11.2q12 (20,490,852-34,117,159 bp) (build 37/hg19). The first probe in the heterodisomic region mapped 34,162,618 bp. Among the 28 STRs spanning the entire long arm of chromosome 14, fourteen markers gave the results for upd(14)mat, whereas the remaining STRs were uninformative. By comparing the proband and maternal alleles, isodisomy was present between D14S261 in 14q11.2 (20,840,388-20,840,704 bp) and D14S275 in 14q12 (26,696,773-26,697,020 bp) markers; the allelic pattern of D14S1040 in 14q12 (32,211,413-32,211,762 bp) could be associated both to isodisomy and to heterodisomy. The rest of the chromosome from D14S70 in 14q13.1 (34,459,194-34,459,447 bp) to D14S1700 in 14q32.33 (105,977,978-105,978,102 bp) was heterodisomic. Segregation analysis of STRs mapping on the other chromosomes confirmed a biparental inheritance (table 2).
Table 2.
The STRs analyzed along with their position (hg19 map)
| Mother | Father | Proband | Result | CGH/SNP | STR locus | Localization | Position |
|---|---|---|---|---|---|---|---|
| 274–303 | 294 | 303 | ID mat | LOH | D14S261 | 14q11.2 | 20,840,388–20,840,704 |
| 100–104 | 102 | 104 | ID mat | LOH | D14S1023 | 14q11.2 | 21,441,901–21,442,220 |
| 135–150 | 129–152 | 150 | ID mat | LOH | D14S283 | 14q11.2 | 22,687,415–22,687,784 |
| 148–150 | 144–150 | 150 | ID mat/biparent | LOH | D14S990 | 14q11.2 | 23,586,268–23,586,632 |
| 203–205 | 207–209 | 203 | ID mat | LOH | D14S972 | 14q11.2 | 24,347,553–24,347,945 |
| 149–151 | 149–155 | 149 | ID mat/biparent | LOH | D14S275 | 14q12 | 26,696,773–26,697,020 |
| 109 | 105–113 | 109 | HD/ID mat | LOH | D14S1040 | 14q12 | 32,211,413–32,211,762 |
| 104–110 | 106–108 | 104–110 | HD mat | normal | D14S70 | 14q13.1 | 34,459,194–34,459,447 |
| 205–213 | 211–213 | 205–213 | HD mat/biparent | normal | D14S75 | 14q13.3 | 37,427,727–37,428,001 |
| 209 | 205–207 | 209 | HD/ID mat | normal | D14S288 | 14q21.2 | 44,101,769–44,102,045 |
| 245–249 | 241–243 | 245–249 | HD mat | normal | D14S276 | 14q22.3 | 55,683,016–55,683,343 |
| 164 | 162–176 | 164 | HD/ID mat | normal | D14S980 | 14q22.3 | 57,152,479–57,152,790 |
| 121 | 121 | 121 | not informative | normal | D14S274 | 14q22.3 | 57,659,338–57,659,723 |
| 187–195 | 185–191 | 187–195 | HD mat | normal | D14S63 | 14q23.2 | 64,651,007–64,651,274 |
| 196–200 | 198–202 | 196–200 | HD mat | normal | D14S258 | 14q24.2 | 70,582,852–70,583,191 |
| 126–128 | 134–136 | 126–128 | HD mat | normal | D14S1036 | 14q24.3 | 75,796,933–75,797,278 |
| 305–309 | 305–307 | 305–309 | HD mat/biparent | normal | D14S74 | 14q24.3 | 78,658,380–78,658,697 |
| 124–126 | 126–138 | 124–126 | HD mat/biparent | normal | D14S1037 | 14q31.3 | 85,197,055–85,197,429 |
| 320–326 | 324–326 | 320–326 | HD mat/biparent | normal | D14S68 | 14q31.3 | 88,627,635–88,627,975 |
| 172 | 172 | 172 | not informative | normal | D14S1044 | 14q32.11 | 90,070,393–90,070,776 |
| 246 | 244–248 | 246 | HD/ID mat | normal | D14S280 | 14q32.12 | 92,182,867–92,183,198 |
| 218–225 | 218–227 | 218–225 | HD mat/biparent | normal | D14S1050 | 14q32.12 | 92,915,524–92,915,918 |
| 162–168 | 164–166 | 162–168 | HD mat | normal | D14S1054 | 14q32.13 | 95,296,491–95,296,837 |
| 148–150 | 144–150 | 148–150 | HD mat/biparent | normal | D14S65 | 14q32.2 | 97,621,472–97,762,169 |
| 248–254 | 250–254 | 248–254 | HD mat/biparent | normal | D14S985 | 14q32.2 | 101,296,536–101,296,815 |
| 226 | 226–233 | 226 | HD/ID mat/biparent | normal | D14S1051 | 14q32.31 | 102,230,242–102,230,439 |
| 91 | 91–93 | 91 | HD/ID mat/biparent | normal | D14S292 | 14q.32.33 | 104,596,704–104,596,962 |
| 91–103 | 103–105 | 91–103 | HD mat/biparent | normal | D14S1007 | 14q32.33 | 105,977,978–105,978,102 |
biparent, a biparental pattern of inheritance; HD, heterodisomy; ID, isodisomy; LOH, loss of heterozygosity; mat, maternal inheritance; STR, short tandem repeat.
Discussion
A de novo robertsonian translocation 45,XY,rob(13;14)(q10;q10) was identified in the proband without chromosomal imbalances by CGH/SNP array. However, a LOH region in 14q11.2q13.1 was found. The final diagnosis of upd(14)mat was performed by microsatellite analysis, which showed a combination of heterodisomy and isodisomy for different regions of chromosome 14. Only employing array CGH would not have revealed heterodisomy, and STR analysis alone would not have defined the LOH extension so accurately. About 50 individuals with altered imprinted gene expression at chromosome 14q32 have been reported [Kotzot, 1999; Hoffmann and Heller, 2011; Ioannides et al., 2014]. In the majority, approximately 75%, upd(14)mat represents the underlying molecular etiology [Ioannides et al., 2014; Briggs et al., 2016]. Patients with Temple syndrome secondary to a paternal deletion at 14q32 or an isolated imprinting defect in the differentially methylated regions on 14q32 have also been reported, both appearing to be of relatively equal frequency [Ioannides et al., 2014; Briggs et al., 2016]. Recently, a single patient with Temple syndrome and multilocus imprinting disturbance has been reported [Bens et al., 2016]. Besides the typical clinical features, this patient showed an abnormal EEG and a pituitary microadenoma [Bens et al., 2016]. So far, there are not enough cases to stratify the clinical findings by (epi)genotypes [Ioannides et al., 2014]. Patients with both hetero- and isodisomic regions are very rare [Antonarakis et al., 1993; Hoffmann and Heller, 2011], but the prevalence of such cases may be underestimated due to the limited number of STRs routinely analyzed during the diagnostic workflow. Albeit alternating segments of heterodisomy and isodisomy should be found in most of upd(14)mat patients as a consequence of meiotic recombination, UPD as the result of robertsonian translocation is rare, approximately 0.6–0.8% [Shaffer, 2006].
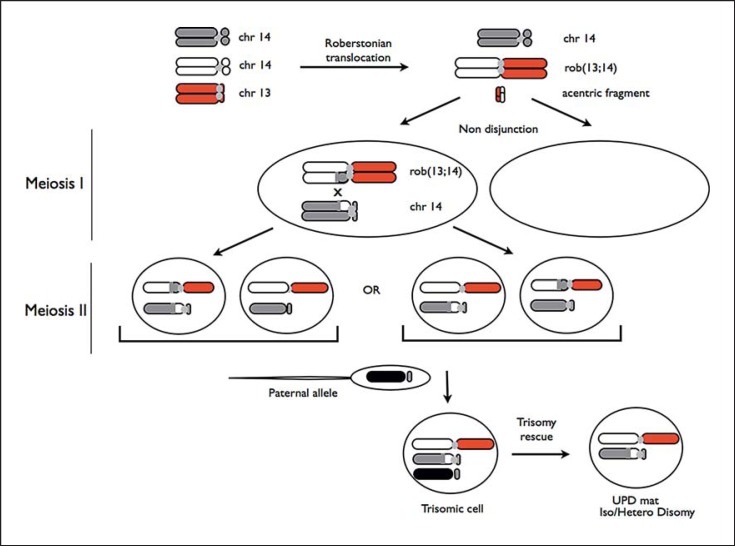
In the present case, a trisomy rescue may be the mechanism involved in the occurrence of upd(14)mat. It can be hypothesized that during maternal meiosis I, 2 pathogenetic events took place: (1) a robertsonian translocation rob(13;14), and (2) a nondisjunction event between 2 chromosomes 14. During maternal meiosis II, the segregation of the chromatids determined the formation of 2 oocytes with a robertsonian translocation t(13;14) and a free chromosome 14 oocyte, which differ in the pattern of UPD (Fig. 2). According to array-CGH results, during maternal meiosis I, a crossing-over occured between chromatides of the homologous chromosomes 14 between 34,117,159 bp and 34,162,618 bp. Since the array-CGH platforms do not usually present oligos in the close proximity of the centromere, we cannot exclude that a second crossing-over has taken place, leading to a heterodisomic region between the centromere and 20,490,852 bp, as described by Antonarakis et al. [1993]. In this hypothetical mechanism, one of these abnormal maternal gametes could then have been fertilized by a normal paternal gamete, leading to a trisomic zygote, followed by a trisomic rescue with the loss of the paternal copy of chromosome 14 [Balbeur et al., 2016]. This mechanism may be characterized by a mosaic trisomy involving the UPD chromosome, as recently demonstrated in a 15-year-old girl [Balbeur et al., 2016]. We did not find a chromosome 14 mosaicism, but a very low level of mosaicism in the blood or its presence in other tissues cannot be excluded.
Fig. 2.
Scheme showing the hypothesized mechanism for the formation of UPD in the proband. In the figure, only the chromosomes involved in the pathological mechanism are shown. The 2 pairs of oocytes with the possible combinations of chromatids in meiosis II are shown.
The isodisomic region can influence the phenotype by unmasking recessive alleles. According to the results of CGH/SNP array, the isodisomic region spans about 13.6 Mb and includes several genes; 24 genes are associated with a well-defined clinical phenotype, and about half of them have a recessive inheritance pattern [www.ncbi.nlm.nih.gov/omim]. Indeed, none of them seem to be strictly related to the phenotypic features in our case (Table 3).
Table 3.
The 24 OMIM genes present in the isodisomic region
| No. | Locus | Position | Gene | Gene name | Phenotype | OMIM | Inheritance |
|---|---|---|---|---|---|---|---|
| 1 | 14q11.2 | 20,684,176–20,694,185 | ANG, RNASE5, ALS9 | angiogenin | amyotrophic lateral sclerosis 9 | 611895 | unknown |
| 2 | 14q11.2 | 21,287,976–21,351,315 | RPGRIP1, LCA6, CORD13 | retinitis pigmentosa GTPase regulator-interacting protein |
Leber congenital amaurosis 6 cone-rod dystrophy 13 |
613826 608194 |
unknown unknown |
| 3 | 14q11.2 | 21,385,193–21,437,297 | CHD8, DUPLIN, KIAA1564, AUTS18 | chromodomain helicase DNA-binding protein 8 | autism, susceptibility to, 18 | 615032 | AD |
| 4 | 14q11.2 | 21,521,079–21,537,215 | SALL2, Hsal2, COLB | spalt-like transcription factor 2, sal like 2 | ?coloboma, ocular, autosomal recessive | 216820 | AR |
| 5 | 14q11.2 | 22,547,505–22,552,131 | TRAC, TRCA, TRA, IMD7 | T-cell receptor alpha | immunodeficiency 7, TCR-alpha/beta deficient | 615387 | AR |
| 6 | 14q11.2 | 22,773,221–22,819,810 | SLC7A7, LPI | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | lysinuric protein intolerance | 222700 | AR |
| 7 | 14q11.2 | 22,836,532–22,847,599 | MMP14, WNCHRS | matrix metalloproteinase 14 (membrane inserted) | ?Winchester syndrome | 277950 | unknown |
| 8 | 14q11.2 | 23,117,305–23,119,610 | CEBPE, CRP1 | CCAAT/enhancer-binding protein (C/EBP), epsilon | specific granule deficiency | 245480 | AR |
| 9 | 14q11.2 | 23,320,187–23,326,184 | PABPN1, PABP2, PAB2 | poly(A)-binding protein, nuclear 1 | oculopharyngeal muscular dystrophy | 164300 | AD |
| 10 | 14q11.2 | 23,381,989–23,408,276 | MYH6, ASD3, MYHCA, CMD1EE, CMH14, SSS3 | myosin heavy chain 6, myosin, heavy polypeptide-6, cardiac muscle, alpha | atrial septal defect 3 sick sinus syndrome 3 cardiomyopathy, hypertrophic, 14 cardiomyopathy, dilated, 1EE |
614089 614090 613251 613252 |
unknown unknown unknown unknown |
| 11 | 14q11.2 | 23,412,737–23,435,685 | MYH7, CMH1, MPD1, CMD1S, SPMM, SPMD | myosin heavy chain 7, myosin, heavy polypeptide-7, cardiac muscle, beta | cardiomyopathy, hypertrophic, 1 scapuloperoneal syndrome, myopathic type cardiomyopathy, dilated, 1S myopathy, myosin storage myopathy, myosin storage Liang distal myopathy left ventricular noncompaction 5 |
192600 181430 613426 255160 608358 160500 613426 |
AD AD AD AR AD AD AD |
| 12 | 14q11q12 | 24,078,692–24,114,923 | NRL, D14S46E, RP27 | neural retina leucine zipper | retinal degeneration, clumped pigment type retinitis pigmentosa 27 |
613750 |
AR AD |
| 13 | 14q11q12 | 24,094,130–24,104,131 | PCK2, PEPCK2 | phosphoenolpyruvate carboxykinase 2, mitochondrial | PEPCK deficiency, mitochondrial | 261650 | AR |
| 14 | 14q12q22 | 24,100,000–57,600,000 | ARVD3 | arrhythmogenic right ventricular dysplasia 3 | arrhythmogenic right ventricular dysplasia 3 | 602086 | AD |
| 15 | 14q12 | 24,100,000–32,900,000 | DFNB5 | deafness, autosomal recessive 5 | deafness, autosomal recessive 5 | 600792 | AR |
| 16 | 14q12q21 | 24,100,000–50,400,000 | SPG32 | spastic paraplegia 32 | spastic paraplegia 32 | 611252 | AR |
| 17 | 14q12 | 24,239,640–24,242,673 | TINF2, TIN2, DKCA3 | TRFl-interacting nuclear factor 2 | Revesz syndrome dyskeratosis congenita, AD 3 |
268130 613990 |
AD AD |
| 18 | 14q12 | 24,249,113–24,263,209 | TGM1, ICR2, ARCI1 | transglutaminase 1, K polypeptide epidermal type I, protein-glutamine gamma-glutamyltransferase | ichthyosis, congenital, AR 1 | 242300 | AR |
| 19 | 14q12 | 28,767,071–28,770,276 | FOXG1, FOXG1B, FKHL1, FKH2, QIN, BF1 | forkhead box G1B | Rett syndrome, congenital variant | 613454 | isolated cases |
| 20 | 14q12 | 30,874,495–30,890,617 | COCH, DFNA9 | cochlin | deafness, autosomal dominant 9 | 601369 | AD |
| 21 | 14q12 | 31,025,105–31,096,449 | AP4S1, CPSQ6, SPG52 | adaptor-related protein complex 4, sigma-1 subunit | spastic paraplegia 52 | 614067 | AR |
| 22 | 14q12 | 31,561,384–31,861,292 | NUBPL, IND1 | nucleotide-binding protein-like protein | mitochondrial complex I deficiency | 252010 | AR, mitochondrial, XL |
| 23 | 14q13 | 32,900,000–37,400,000 | HPE8 | holoprosencephaly 8 | holoprosencephaly 8 | 609408 | unknown |
| 24 | 14q13q21 | 32,900,000–50,400,000 | RLS2 | restless legs syndrome, susceptibility to, 2 | {restless legs syndrome 2} | 608831 | unknown |
AD, autosomal dominant; AR, autosomal recessive; XL, X-linked dominant. Genomic coordinate from NCBI/GRCh38.
In the literature, the phenotype associated with upd(14)mat is usually mild with a relevant degree of variability as summarized in Table 4. Frequent features, such as pre- and postnatal growth retardation, psychomotor delay, facial dysmorphisms, and short hands and feet, are present in our proband (Table 4). He also shares mild neurodevelopmental disability with upd(14)mat (Table 4). In addition, truncal obesity developed after initial failure to thrive in the first months of life due to feeding problems. He did not manifest the compulsive eating habits typically seen in patients with Prader-Willy syndrome [Butler et al., 2016]. This aspect can be helpful for clinical differential diagnosis in addition to the other phenotypic features [Hoffmann and Heller, 2011]. The boy also showed bilateral undescended testes which is very rarely reported in this syndrome [Ioannides et al., 2014], while precocious onset of puberty, which is a frequent feature (Table 4), was not present, likely due to the young age at the last evaluation. The boy presented with hypoesthesia to pain, an unreported finding in individuals with Temple syndrome [Ioannides et al., 2014], but additional observations are needed to confirm this finding on the clinical spectrum of upd(14)mat. Moreover, the phenotypical features usually unreported in patients with Temple syndrome could be caused by a recessive gene mutation in the isodisomic region. Several of the above-mentioned findings may suggest a hypothalamic-pituitary dysfunction, but functional and imaging investigations of this area were not performed in our case as well as in the majority of the previous reported patients [Ioannides et al., 2014].
Table 4.
Major clinical findings of the proband and individuals with Temple syndrome due to upd(14)mat
| Proband | Literature dataa, % | |
|---|---|---|
| Physical features | ||
| Premature birth | – | 40 |
| Intrauterine growth retardation | + | 79 |
| Low birth length (<5th centile)b | + | 55 |
| Low birth weight (<5th centile)b | + | 86 |
| Head circumference at birth (<5th centile)b | – | 28 |
| Postnatal short stature (<5th centile)b | + | 81 |
| Small hands | + | 83 |
| Small feet | + | 95 |
| Truncal obesity | + | 50 |
| Precocious/early puberty | – | 87 |
| Neurological and musculoskeletal findings | ||
| Hypotonia | + | 91 |
| Feeding problems (infants) | + | 16c |
| Joint hypermobility | + | 60 |
| Scoliosis | – | 26 |
| Motor developmental delay | + | 81 |
| Speech delay | + | 45 |
| Intellectual disability | + | 42 |
Ioannides et al. [2014].
Limited to the first months of life, with progressive spontaneous improvement thereafter.
Raw number of patients.
Conclusions
Clinical diagnosis of upd(14)mat or Temple syndrome is usually difficult at birth and in early childhood, since the typical clinical findings (i.e., truncal obesity, precocious puberty, and adult short stature) are not yet present. If a conventional cytogenetic analysis is done on the background of mild phenotypic features, in the presence of a robertsonian translocation, the STR analysis, spanning the entire chromosome 14, is mandatory. This test represents the gold standard to reveal the UPD and to detect hetero- or isodisomy (13;14). The CGH/SNP array should always flank STR analysis in order to exclude genomic imbalances, and to better define the extent of LOH, where recessive disease alleles can be unmasked and contribute to characterize the pathological phenotype. Early genetic diagnosis permits a specific follow-up, including rehabilitative neurodevelopmental programs and preventive efforts regarding the management of obesity, precocious puberty, and short stature in order to optimize the long-term outcome. Uncertainty persists regarding hypothalamic-pituitary dysregulation, and it should be assessed in future studies.
Statement of Ethics
The study was conducted according to the Declaration of Helsinki and the standard protocol of investigation of a child with obesity and neurodevelopmental disability in our Departments. The parents had given their informed written consent before any clinical and genetic investigation.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- Antonarakis SE, Blouin JL, Maher J, Avramopoulos D, Thomas G, Talbot CC., Jr Maternal uniparental disomy for human chromosome 14, due to loss of a chromosome 14 from somatic cells with t(13;14) trisomy 14. Am J Hum Genet. 1993;52:1145–1152. [PMC free article] [PubMed] [Google Scholar]
- Balbeur S, Grisart B, Parmentier B, Sartenaer D, Leonard PE, et al. Trisomy rescue mechanism: the case of concomitant mosaic trisomy 14 and maternal uniparental disomy 14 in a 15-year-old girl. Clin Case Rep. 2016;4:265–271. doi: 10.1002/ccr3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bens S, Kolarova J, Beygo J, Buiting K, Caliebe A, et al. Phenotypic spectrum and extent of DNA methylation defects associated with multilocus imprinting disturbances. Epigenomics. 2016;8:801–816. doi: 10.2217/epi-2016-0007. [DOI] [PubMed] [Google Scholar]
- Bertino E, Spada E, Occhi L, Coscia A, Giuliani F, et al. Neonatal anthropometric charts: the Italian neonatal study compared with other European studies. J Pediatr Gastroenterol Nutr. 2010;51:353–361. doi: 10.1097/MPG.0b013e3181da213e. [DOI] [PubMed] [Google Scholar]
- Beygo J, Elbracht M, de Groot K, Begemann M, Kanber D, et al. Novel deletions affecting the MEG3-DMR provide further evidence for a hierarchical regulation of imprinting in 14q32. Eur J Hum Genet. 2015;23:180–188. doi: 10.1038/ejhg.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs TA, Lokulo-Sodipe K, Chandler KE, Mackay DJG, Temple IK. Temple syndrome as a result of isolated hypomethylation of the 14q32 imprinted DLK1/MEG3 region. Am J Med Genet A. 2016;170A:170–175. doi: 10.1002/ajmg.a.37400. [DOI] [PubMed] [Google Scholar]
- Buiting K, Kanber D, Martín-Subero JI, Lieb W, Terhal P, et al. Clinical features of maternal uniparental disomy 14 in patients with an epimutation and a deletion of the imprinted DLK1/GTL2 gene cluster. Hum Mutat. 2008;29:1141–1146. doi: 10.1002/humu.20771. [DOI] [PubMed] [Google Scholar]
- Butler MG, Manzardo AM, Forster JL. Prader-Willi syndrome: clinical genetics and diagnostic aspects with treatment approaches. Curr Pediatr Rev. 2016;12:136–166. doi: 10.2174/1573396312666151123115250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6:137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Heller R. Uniparental disomies 7 and 14. Best Pract Res Clin Endocrinol Metab. 2011;25:77–100. doi: 10.1016/j.beem.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Ioannides Y, Lokulo-Sodipe K, Mackay DJ, Davies JH, Temple IK. Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J Med Genet. 2014;51:495–501. doi: 10.1136/jmedgenet-2014-102396. [DOI] [PubMed] [Google Scholar]
- Kagami M, Matsuoka K, Nagai T, Yamanaka M, Kurosawa K, et al. Paternal uniparental disomy 14 and related disorders: placental gene expression analyses and histological examinations. Epigenetics. 2012;7:1142–1150. doi: 10.4161/epi.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzot D. Abnormal phenotypes in uniparental disomy (UPD): fundamental aspects and a critical review with bibliography of UPD other than 15. Am J Med Genet. 1999;82:265–274. [PubMed] [Google Scholar]
- Ogata T, Kagami M. Kagami-Ogata syndrome: a clinically recognizable upd(14)pat and related disorder affecting the chromosome 14q32.2 imprinted region. J Hum Genet. 2016;61:87–94. doi: 10.1038/jhg.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays. 2000;22:452–459. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Shaffer LG. Risk estimates for uniparental disomy following prenatal detection of a nonhomologous Robertsonian translocation. Prenat Diagn. 2006;26:303–307. doi: 10.1002/pd.1384. [DOI] [PubMed] [Google Scholar]
- Tanner JM, Whitehouse RH. Clinical longitudinal standards for height, weight, height velocity, weight velocity, and stages of puberty. Arch Dis Child. 1976;51:170–179. doi: 10.1136/adc.51.3.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple IK, Shrubb V, Lever M, Bullman H, Mackay DJ. Isolated imprinting mutation of the DLK1/GTL2 locus associated with a clinical presentation of maternal uniparental disomy of chromosome 14. J Med Genet. 2007;44:637–640. doi: 10.1136/jmg.2007.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]