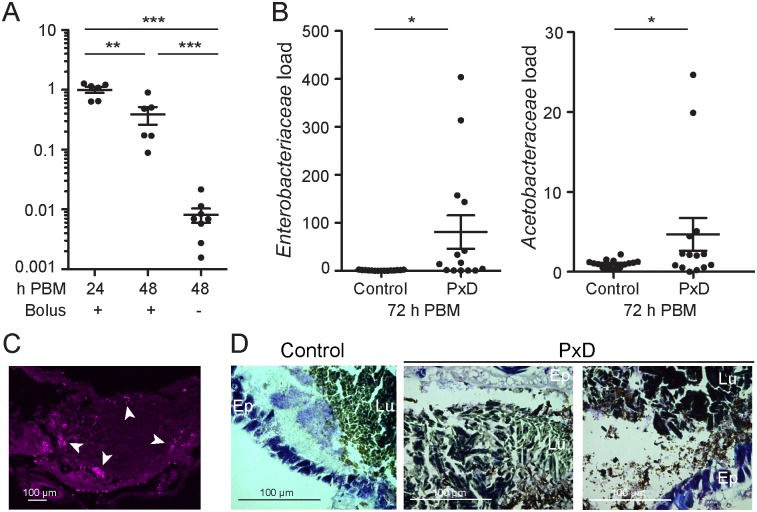
Fig 4. The peritrophic matrix promotes restoration of gut homeostasis after blood feeding.
(A) Midgut bacterial load at 24h and 48h after a blood meal, as determined by qRT-PCR. At 48h, midguts were pooled according to whether (+) or not (-) the blood bolus was present. Each dot represents a pool of 5 guts, derived from two independent experiments. Ratios are normalized within biological replicates to the mean of the 24h pools. Mean plus/minus standard error is indicated. Statistical significance was assessed by an ANOVA on a linear mixed effect regression model. ‘*’ p<0.05; ‘**’ p<0.01; ‘***’ p<0.001. (B) Enterobacteriaceae and Acetobacteraceae load in midguts 72h after feeding on blood supplemented with 100μM polyoxin D (PxD) or an equal volume of water (control), as determined by qRT-PCR. Each dot represents a pool of 8–10 guts, derived from 4 independent experiments. Ratios are normalized within biological replicates to the mean of the control pools. Mean plus/minus standard error is indicated. Statistical significance was assessed and is presented as described above. (C) Thin abdominal section 24h post blood meal stained with anti-LPS antibody. White arrowheads indicate LPS staining. (D) Gram-stained thin abdominal sections 24h post blood meal, with or without polyoxin D supplementation. Bacteria are stained light purple. “Lu” lumen; “Ep” epithelium.