Figure 2.
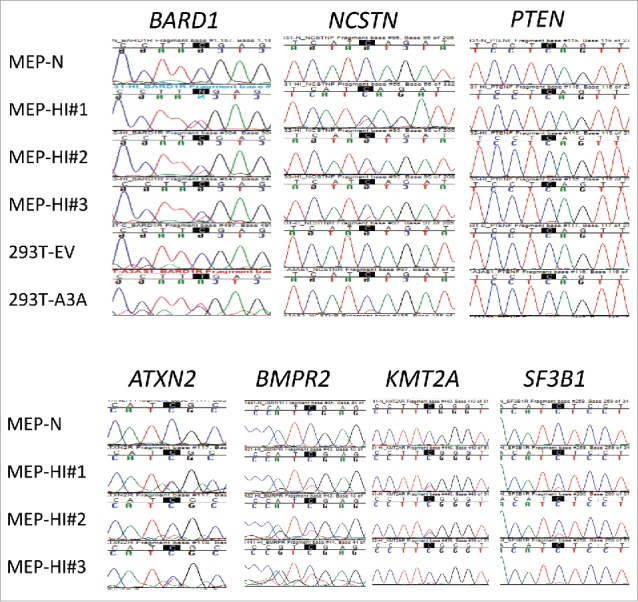
Sanger validation of C > U RNA editing sites in primary cells identified by low-stringency RNA seq analysis of 293T/A3A cells. RNA editing at the highlighted Cs was validated in monocyte/enriched PBMCs (MEPs; n = 3 donors) treated with hypoxia and IFN1 (HI) for 24 h, in 7 of 19 tested genes. C > T(U) editing is characterized by emergence of a secondary T peak (red) accompanied by a reduction in height of C peak (blue). MEP-N shows lack of editing in a representative sequence chromatogram from control MEP cells maintained in normoxia without IFN1 treatment. Chromatograms for the remaining 12 RNA editing sites that could not be validated in MEPs are shown Fig. S2.
