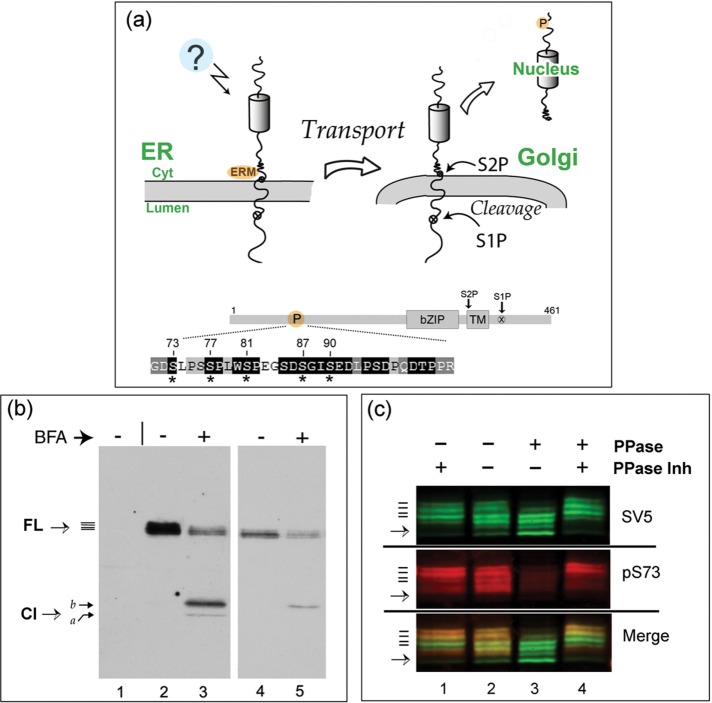
FIGURE 1:
FL CREB-H is multiply phosphorylated within the P-motif. (a) Diagram illustrating the precursor form of CREB-H anchored in the ER membrane. Unidentified signals (indicated by a question mark) induce release from the ER, transport to the Golgi, and subsequent processing by S1P and S2P proteases. Bottom, subdomains of CREB-H: bZIP, the basic-zipper DNA-binding domain; TM, the transmembrane domain; and the S1P and S2P sites. P, the P-motif, with the amino acid sequence expanded below. Residues that are very highly conserved across all CREB-H homologues are indicated by white lettering on a black background (Barbosa et al., 2015). Key serine residues are indicated by asterisks. (b) COS cells were transfected with the expression vector for FL CREB-H and treated without or with BFA (final concentration 1 µg/ml, added 1 h before harvesting). Total cell extracts were analyzed by SDS–PAGE and Western blotting. A mock-transfected control sample is shown in lane 1. Lanes 4 and 5 show a shorter exposure of lanes 2 and 3. CREB-H FL precursor migrates as multiple species (solid bars), as discussed in the text. (c) Dephosphorylation of CREB-H. Soluble extracts in nondenaturing buffer from cells expressing the precursor form of CREB-H were incubated at 37°C with (lane 1) or without phosphatase inhibitors (lane 2) or with added λ-phosphatase again with (lane 4) or without phosphatase inhibitors (lane 3). The total population of CREB-H was detected using the anti-SV5 mouse primary antibody, and phosphorylation of serine 73 was simultaneously detected using the specific anti-pS73 antibody (Barbosa et al., 2015). CREB-H species were simultaneously detected by appropriate fluorochrome- labeled anti-mouse and anti-rabbit secondary antibodies using a LI-COR laser scanning system as described in Materials and Methods. Individual channels are shown separately, as well as together in the merged channel.